3063
The ERIC phantom - Extra-dimensional respiration and inflow of contrast: an open-source DCE digital simulation tool1Radiology and Nuclear Medicine, Academic Medical Centre, Amsterdam, Netherlands
Synopsis
Keywords: Software Tools, DSC & DCE Perfusion
An open-source MATLAB-based software tool was developed for digital simulation (using the MRXCAT framework) of DCE MRI acquisition and reconstruction strategies, termed the ERIC phantom: extra-dimensional respiration and inflow of contrast. The ERIC phantom is a user-friendly graphical user interface, allowing for variable respiratory motion, acquisition parameters and trajectories, motion correction strategies, and compressed sensing reconstructions. This realistic abdominal DCE phantom can be used to investigate varying strategies for producing high quality DCE reconstructions in the presence of respiratory motion and contrast inflow.Background
Dynamic contrast-enhanced (DCE) MRI is widely used to estimate tissue perfusion parameters. In abdominal DCE, sampling and reconstructing of 3D volumes which cover the whole abdomen over short (≈3 second) DCE frames is hindered by respiratory motion and data sampling constraints. This is particularly challenging in organs like the pancreas, which is elongated, small (relative to e.g. kidneys and liver), and centrally located.These factors have led previous research to utilize fast MR undersampling strategies (e.g. keyhole imaging1, stack-of-stars/spirals2,3, variable density Cartesian4) coupled with motion mitigation (e.g. respiratory soft-gating4, auto-focus motion correction5) and iterative reconstruction algorithms (e.g. compressed sensing2,6).
One remaining challenge in abdominal DCE MRI is the inherent lack of ground truth data and the burden of scanning patients and healthy controls with contrast injection. Thus optimal scan and reconstruction settings are difficult to establish. Digital phantoms allow for interactive and quick testing, and may be elucidative in the optimization of scan acquisition and reconstruction protocols. For a digital phantom to be useful, it should be open-source, well-documented, easy-to-use, and, most importantly, able to generate results similar to those gained from in-vivo experiments.
This work aims to provide a user-friendly MATLAB-based app to interactively explore a DCE MRI digital phantom, based on the MRXCAT implementation7, entitled ERIC: Extra-dimensional Respiration and Inflow of Contrast. This open-source and freely-available tool (https://github.com/schrau24/XCAT-ERIC) permits flexible simulation, sampling, and reconstruction parameter testing to optimize a given application for abdominal DCE MRI.
Workflow
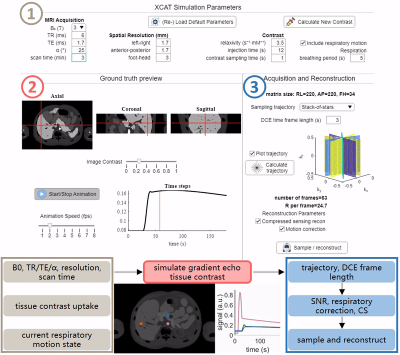
See Figure 1 for an overview. The app was designed in MATLAB 2021a (Mathworks, Natick, MA, USA) on a Windows PC with 32 GB RAM and an Intel-Neon® E5-1620 processor (3.40 GHz, 4 cores).The ERIC phantom enables full choice of sequence parameters: B0, TR, TE, α, and spatial resolution. For the user-defined coverage of the abdomen, these parameters are used in the spoiled GRE signal equation per tissue to generate realistic image contrast. Breathing motion is induced through the user choice of respiratory period length – here, expiration is assumed to be ≈40% of the respiratory period. Dynamic inflow of gadolinium-based contrast in the liver, spleen, and pancreas is simulated. Signal curves in each organ are derived from previously published DCE parameters using the extended Tofts model8,9. Finally, the 4D ground truth images are presented, allowing for interactive interrogation of contrast inflow and respiration effects per tissue.
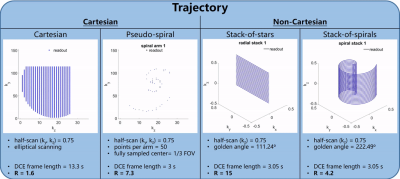
Following the generation of simulated ground truth images, an acquisition trajectory is selected, calculated for the duration of the scan, and sorted for the defined reconstructed DCE time frame length. Feedback for plotted trajectories, number of reconstructed DCE frames, and undersampling factor R is shown. See Figure 2 for example trajectory calculations, display, and feedback (B0 = 3T, scan time = 3 min, TR/TE = 5.1/1.7 ms, α = 25°, matrix = 220 x 152 x 35, target DCE frame length = 3 s). Spiral samples are calculated using a rapid center-out variable density scheme10.
Data sampling using the prescribed trajectory is performed for each unique combined contrast and respiratory phase. For Cartesian trajectories, k-space from the current image is produced through simple 3D FFT and individual readouts from to the current sampling time are added to the sampled set. With non-Cartesian trajectories a true 3D non-uniform FFT (NUFFT, https://github.com/marcsous/nufft_3d) is used for sampling. Respiratory compensation or correction can be chosen to be either a soft-gating4 or autofocus approach5.
DCE CS reconstructions are performed utilizing established techniques for comparison of resulting DCE images using bart6 (Cartesian) or GRASP2 (non-Cartesian).
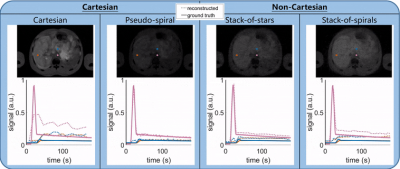
Final simulated reconstructed images and inflow curves can be interactively probed and compared against ground truth inputs and saved for later interrogation. See Figure 3.
Demonstration
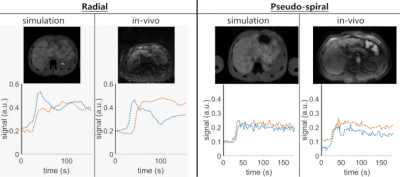
As a demonstration, images from two clinical in-vivo DCE acquisitions are included for comparison against simulation: 1. radial stack-of-stars and 2. pseudo-spiral. In-vivo data were acquired on a 3T MRI system (Ingenia, Philips, Best, the Netherlands) with the parameters – 1. radial stack-of-stars: TR/TE = 3.5/1.4 ms, α = 22°, matrix = 232 x 232 x 66, resolution = 1.5 x 1.5 x 3.5 mm, slice half-scan = 0.75, golden angle = 111.2°, reconstructed DCE frame length = 6 s, and resulting R = 18.2; 2. pseudo-spiral: TR/TE = 5.1/1.7 ms, α = 25°, matrix = 220 x 152 x 34, resolution = 1.7 x 1.7 x 3 mm, phase/slice half-scan = 0.75, reconstructed DCE frame length = 3 s, and resulting R = 7.3. Simulations used matching acquisition and reconstruction parameters, including acceleration factor R and simulated respiration motion chosen to reflect what was observed in-vivo. See Figure 4 for results.Acknowledgements
No acknowledgement found.References
1. Raja, R. & Sinha, N. Adaptive k-space sampling design for edge-enhanced DCE-MRI using compressed sensing. Magn. Reson. Imaging 32, 899–912 (2014).
2. Feng, L. et al. Golden-angle radial sparse parallel MRI: combination of compressed sensing, parallel imaging, and golden-angle radial sampling for fast and flexible dynamic volumetric MRI. Magn. Reson. Med. 72, 707–717 (2014).
3. Weller, D. S., Wang, L., Mugler, J. P. & Meyer, C. H. Motion-Compensated Reconstruction of Magnetic Resonance Images from Undersampled Data. Magn. Reson. Imaging 55, 36 (2019).
4. Cheng, J. Y. et al. Free-breathing pediatric MRI with nonrigid motion correction and acceleration. J. Magn. Reson. Imaging 42, 407–420 (2015).
5. Atkinson, D., Hill, D. L. G., Stoyle, P. N. R., Summers, P. E. & Keevil, S. F. An autofocus algorithm for the automatic correction of motion artifacts in MR images. Lect. Notes Comput. Sci. (including Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinformatics) 1230, 341–354 (1997).
6. Uecker, M. & Tamir, J. mrirecon/bart: version 0.5.00. (2019) doi:10.5281/ZENODO.3376744.
7. Wissmann, L., Santelli, C., Segars, W. P. & Kozerke, S. MRXCAT: Realistic numerical phantoms for cardiovascular magnetic resonance. J. Cardiovasc. Magn. Reson. 16, 1–11 (2014).
8. Klaassen, R. et al. Repeatability and correlations of dynamic contrast enhanced and T2* MRI in patients with advanced pancreatic ductal adenocarcinoma. Magn. Reson. Imaging 50, 1–9 (2018).
9. Holland, M. D. et al. Disposable point‐of‐care portable perfusion phantom for quantitative DCE‐MRI. Med. Phys. 49, 271–281 (2022).
10. Lee, J. H., Hargreaves, B. A., Hu, B. S. & Nishimura, D. G. Fast 3D Imaging Using Variable-Density Spiral Trajectories with Applications to Limb Perfusion. Magn. Reson. Med. 50, 1276–1285 (2003).
11. Lo, W. et al. Realistic 4D MRI abdominal phantom for the evaluation and comparison of acquisition and reconstruction techniques. Magn. Reson. Med. 81, 1863–1875 (2019).
Figures