3059
Relative Cerebral Blood Volume Differences Between Adult-Type Diffuse Glioma Subgroups According to WHO 2021: A Study of 146 Gliomas1Institute of Biomedical Engineering, Bogazici University, Istanbul, Turkey, 2Department of Medical Pathology, Acibadem University, Istanbul, Turkey, 3Brain Tumor Research Group, Acibadem University, Istanbul, Turkey, 4Department of Molecular Biology and Genetics, Acibadem University, Istanbul, Turkey, 5Department of Neurosurgery, Acibadem University, Istanbul, Turkey, 6Department of Radiology, Acibadem University, Istanbul, Turkey
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Machine Learning/Artificial Intelligence
The main purpose of this study is to analyze the relative cerebral blood volume (rCBV) differences of adult diffuse glioma subgroups defined in the updated WHO 2021 brain tumor classification. The IDH wildtype group had statistically significantly higher rCBV values, and IDH mutational subgroups were classified with 82.5% accuracy (precision = 82.1%, recall = 82.9%), while the accuracies of glioblastoma and oligodendroglioma classification was 81.9%, glioblastoma and astrocytoma classification was 83.3%, and astrocytoma and oligodendroglioma classification was 77.5%.Introduction
Isocitrate dehydrogenase (IDH) mutation highly affects the prognosis and clinical outcome in gliomas1,2. According to WHO 2021 update, IDH wild type (IDH-wt) gliomas are classified as glioblastoma, regardless of their grade. In addition, astrocytoma and oligodendroglioma in the IDH-mutant (IDH-mut) group are separated according to the 1p19q codeletion status3. Recent studies have associated IDH mutation with tumor angiogenesis in gliomas and reported that the rCBV maps obtained from dynamic susceptibility contrast (DSC) MRI could be used to predict different glioma subgroups4. Therefore, this study aims to analyze the rCBV differences between the diffuse adult glioma subgroups defined in WHO 2021 brain tumor classification guidelines using pixel-wise analysis followed by supervised machine learning algorithms.Methods
A patient cohort of 146 gliomas (55F/91M, mean age = 47.1 ± 14.5 years, 83 IDH-wt (glioblastoma), 63 IDH-mut including 40 astrocytoma and 23 oligodendroglioma) were included in this study. The patients were scanned on a 3T Siemens MR scanner (Erlangen, Germany) using a 32-channel head coil before surgery. The brain tumor protocol included pre- and post-contrast (gadolinium DTPA) T1-weighted TSE (TR=500 ms, TE=10 ms), T2-weighted TSE (TR=5000 ms, TE=105 ms), and T2*-weighted gradient-echo echo-planar imaging (EPI) DSC-MRI (TR=1500 ms, TE=30 ms). The whole tumor volumes were segmented on FLAIR images, and the necrosis volumes were segmented on T1-weighted post-contrast images using 3D slicer version 4.8.1 (http://slicer.org/). Each subject's tumor and necrosis areas were registered to the rCBV maps using ANTs5. The histogram properties such as median, mean, standard deviation, kurtosis, and skewness of the rCBV pixel values within the tumor regions were calculated using an in-house program written in MATLAB 2020a (The MathWorks Inc., Natick, MA). A Kruskal-Wallis test was used with multiple comparisons for assessing rCBV feature differences between the astrocytoma, oligodendroglioma, and glioblastoma subgroups. Since the dataset size is relatively small, a power analysis was also performed. Then, different histogram properties were given as input features to supervised machine learning algorithms with 10-fold cross-validation using an in-house Python script.Results
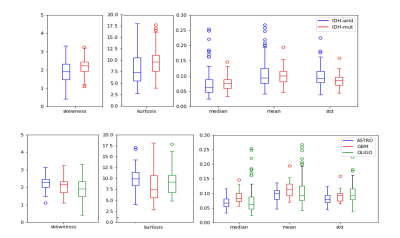
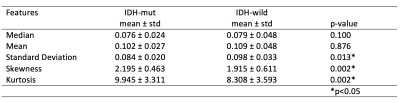
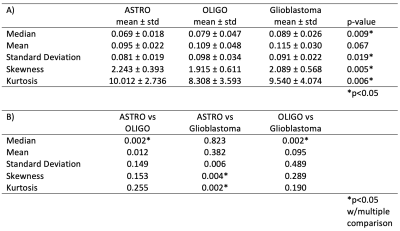
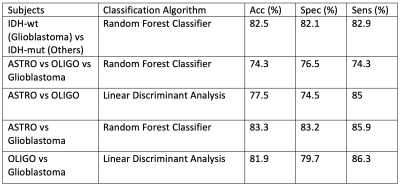
The standard deviation, skewness, and kurtosis of the rCBV values were statistically significantly different between IDH-mut and IDT-wt tumors (p<0.05) (Table 1). The power analysis was also performed for this group, which was higher than 80%. Moreover, IDH-wt tumors had higher mean values than the IDH-mut ones (Figure 1). Glioblastomas had higher median values than oligodendrogliomas and lower skewness and kurtosis than astrocytomas (Figure 1). On the other hand, oligodendrogliomas had higher median values than astrocytomas (Table 2). The power of the Kruskal Wallis test was also higher than 80% for comparing these subgroups.According to the machine learning results, the IDH mutational subgroups were classified with 82.5% accuracy (precision = 82.1%, recall = 82.9%). The accuracy for the 3-class classification task was 74.3% (precision = 76.5%, recall = 74.3%). Moreover, glioblastoma and oligodendrogliomas were classified with 81.9% accuracy (precision = 79.7%, recall = 86.3%), glioblastoma and astrocytomas were classified with 83.3% accuracy (precision = 83.2%, recall = 85.9%), and astrocytomas and oligodendrogliomas were classified with 77.5% accuracy (precision = 74.5%, recall = 85%) (Table 3).
Conclusion and Discussion
The cerebral blood volume differences between updated WHO 2021 adult glioma subgroups were statistically significant. Higher rCBV values and, therefore, higher angiogenesis were observed in the IDH-wt group. In addition, the astrocytoma group was found to have lower rCBV values. Future studies will verify these results on larger datasets.Acknowledgements
This study was supported by the Scientific and Technological Research Council of Turkey (TUBITAK) grant 216S432.References
1. Network CGAR. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. New England Journal of Medicine 2015;372(26):2481-98.
2. Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H, et al. Glioma Groups Based on 1p/19q, IDH, and TERT Promoter Mutations in Tumors. N Engl J Med 2015;372(26):2499-508.
3. Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol 2021;23(8):1231-51.
4. Kickingereder P, Sahm F, Radbruch A, Wick W, Heiland S, Von Deimling A, et al. IDH mutation status is associated with a distinct hypoxia/angiogenesis transcriptome signature which is non-invasively predictable with rCBV imaging in human glioma. Scientific reports 2015;5:16238.
5. Avants BB, Tustison NJ, Stauffer M, Song G, Wu B, Gee JC. The Insight ToolKit image registration framework. Front Neuroinform 2014;8:44.
Figures