3053
Multimodal deep learning for Alzheimer’s disease classification and clinical score prediction1Neuroscience, Emory University, Decatur, GA, United States, 2Georgia Tech, Decatur, GA, United States, 3Georgia State University, Decatur, GA, United States
Synopsis
Keywords: Alzheimer's Disease, Multimodal, Neurodegeneration, Deep Learning
In this project, we use different neuroimaging modalities in the ADNI data, including structural magnetic resonance imaging (sMRI) and temporal transformations of resting-state functional magnetic resonance imaging (rs-fMRI) data, for several classification (e.g., diagnosis) and regression (e.g., age and clinical assessments) objectives.Introduction
Alzheimer’s disease is a neurodegenerative and progressive disease. The Alzheimer’s Disease Neuroimaging Initiative (ADNI) is a robust study to acquire imaging, clinical, genetic, and several other biomarkers of individuals ranging from mild cognitive impairment (MCI) to dementia. In this project, we use different neuroimaging modalities in the ADNI data, including structural magnetic resonance imaging (sMRI) and temporal transformations of resting-state functional magnetic resonance imaging (rs-fMRI) data, for several classification (e.g., diagnosis) and regression (e.g., age and clinical assessments) objectives. We did several temporal transformations such as the amplitude of low-frequency fluctuations (ALFF), the fractional amplitude of low-frequency fluctuations (fALFF), Kendall's coefficient of concordance Regional Homogeneity (KccReHo), voxel-mirrored homotopic connectivity (VMHC), degree centrality (DC), Percent Amplitude of fluctuation (PerAF), which have been shown to contribute towards data reduction while preserving the voxel dimensionality1.Methods
We used the structural (sMRI) and resting-state functional (fMRI) magnetic resonance imaging scans from the Alzheimer’s Disease Neuroimaging Initiative (ADNI)3. The T1-weighted sMRI scans were preprocessed using Statistical Parametric Mapping (SPM) Toolbox 12. We did segmentation of the sMRI images to isolate gray matter brain areas using MNI template and further smoothed (using a 3D Gaussian kernel), modulated and warped to extract the gray matter2. The fMRI scans were further processed using temporal transformations. We used a 3D Convolutional Neural Network - AlexNet2 for classification and regression tasks. The performance of the classification model was assessed using test accuracy whereas regression tasks were assessed using mean absolute error (mae) and coefficient of determination/R-squared (r2). 10 iterations were performed for each task and mean and standard deviation were computed and plotted on the graphs (Figure 1 and Figure 2).Results
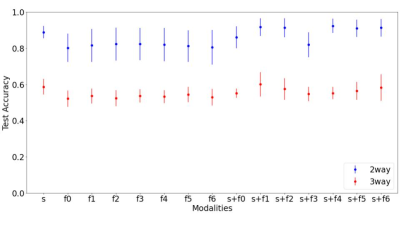
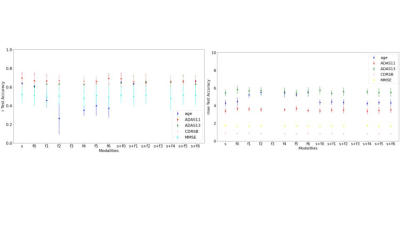
Figure 1 indicates the classification results and we observe that sMRI unimodal outperforms all the fMRI unimodal results. We do observe some multimodal models with temporal features (two way - KccReHo, Degree Centrality,VMHC, fALFF; three-way - KccReHo) and sMRI perform either equivalently or better than the unimodal models for classification of both two-ways (AD vs CN) and three-ways (AD vs CN vs MCI). We also observe that all multimodal models perform better than their corresponding unimodal fMRI models. Figure 2 indicates the regression results where we see that for ADAS11 and MMSE, the multimodal models (with temporal transformation Degree Centrality - Positive Binarised Sum and Degree Centrality - Positive Weighted Sum) shows the best results. For other regression scores ADAS13, age and CDSR, we find unimodal fMRI models to perform the best.Discussion and Conclusion
The multimodal analyses in this work employs a convolutional neural network (CNN) model to study the unique views of the brain's structural and functional organization presented by the different data modalities. Furthermore, to access the complimentary, joint information in the multimodal data, we propose data fusion4 with a multi-channel CNN for the multimodal analyses in this work. The results from our experiments substantiate the T1-weighted sMRI to be generally the most predictive in all tasks, consistently outperforming the rs-fMRI temporal transformations by 3-7%. We also note a performance gain in our multimodal analyses, reflecting the high potential for multimodal data fusion using deep learning5 architectures to predict diagnosis and clinical assessments. Future work involves studying disease progression and evaluating the spatial distributions of most predictive brain regions for the undertaken tasks via saliency mapping.Acknowledgements
No acknowledgement found.References
1. A. Abrol, R. Hassanzadeh, S. Plis and V. Calhoun, "Deep learning in resting-state fMRI," 2021 43rd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), 2021, pp. 3965-3969, doi: 10.1109/EMBC46164.2021.9630257.
2. Abrol, A., Fu, Z., Salman, M., Silva, R., Du, Y., Plis, S., & Calhoun, V. (2021). Deep learning encodes robust discriminative neuroimaging representations to outperform standard machine learning. Nature communications, 12(1), 1-17.
3. Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, Harvey D, Jack CR, Jagust W, Liu E, Morris JC, Petersen RC, Saykin AJ, Schmidt ME, Shaw L, Siuciak JA, Soares H, Toga AW, Trojanowski JQ; Alzheimer’s Disease Neuroimaging Initiative. The Alzheimer's Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement. 2012 Feb;8(1 Suppl):S1-68. doi: 10.1016/j.jalz.2011.09.172. Epub 2011 Nov 2. PMID: 22047634; PMCID: PMC3329969.
4. Calhoun, V. D., & Sui, J. (2016). Multimodal fusion of brain imaging data: a key to finding the missing link (s) in complex mental illness. Biological psychiatry: cognitive neuroscience and neuroimaging, 1(3), 230-244.
5. Ramachandram, D., & Taylor, G. W. (2017). Deep multimodal learning: A survey on recent advances and trends. IEEE signal processing magazine, 34(6), 96-108.
Figures