3025
A step forward to improve large numerical phantoms at the microstructure level using CACTUS1Signal Processing Laboratory (LTS5), École polytechnique fédérale de Lausanne, Lausanne, Switzerland, 2Department of Computer Science, Université de Sherbrooke, Monreal, QC, Canada, 3Radiology Department, Centre Hospitalier Universitaire Vaudois and University of Lausanne, Lausanne, Switzerland, 4CIBM Center for Biomedical Imaging, Lausanne, Switzerland
Synopsis
Keywords: White Matter, Phantoms, Monte Carlo simulations for microstructure in DW-MRI
Inferring microscopic tissue properties from the measured signal is one of the main objectives of diffusion-weighted magnetic resonance imaging. The use of Monte-Carlo Simulations for DW-MRI on realistic substrates will help the study of DWI-MRI signals in controlled environments and investigate, extract, and validate approaches for the understanding of white matter features. This work provides a unique framework for building complex white matter phantoms with novel properties, such as 1) substrates with a high packing density of 95% intra-axonal volume percentage and substrate sizes of (500 um)3, 2) Create, generalise, extend, and preexisting phantom configurations used by other studies.Introduction
Monte Carlo simulations are invaluable for analysing microstructure models[9] of Diffusion-Weighted Magnetic Resonance Imaging (DW-MRI). They are critical for examining diffusion phenomena in complex substrates with no analytical solutions, such as the diffusion process in the extracellular space and axons with undulations, variable diameter, and local microdispersion. Contrary to conventional techniques, Monte Carlo simulations don't need an explicit description of the diffusion signal. Instead, it uses a three-dimensional (3D) mesh, a geometric representation of the substrate. A detailed substrate will therefore produce a DW-MRI signal with relevant properties. The challenge of producing such complex white matter substrates has been addressed in previous studies [3,4,8]. However, reaching a desired high axonal packing density in large volumetric phantoms remains an open challenge [7,19].This abstract presents the improvements made to CACTUS (Computational Axonal Configurations for Tailored and Ultradense Substrates), a novel framework with multiple parameters to create substrates mimicking various white matter features. This work describes a novel application of the CACTUS framework for generating complex synthetic substrates. Firstly, we show synthetic substrates with axonal packing of unprecedented intracellular volume fractions (ICVF) and substrate volume of (500 um)3, large enough to represent the tissue properties of interest [7]. Moreover, we show the ability of our framework to create and enhance phantoms inspired by other studies, such as the DiSCo challenge synthetic phantom [12].
Methods
We designed a novel substrate generator with parameterisable white matter features. The creation of the substrate is divided into three steps, shown in Figure 1. Firstly, the fibre initialisation step, where we select the prior distributions of the fibre trajectories and radii used in the substrates. In our case of study, we work with two examples: 1) Groups of fibres arranged in a single fibre population, with a 15º of dispersion w.r.t the main bundle orientation as in NODDI[13] 2) Fibres trajectories spired by the DiSCo challenge[12], where fibres from different white matter configurations (e.g., kissing, branching) intersect at different crossing angles, interconnecting 16 Regions of Interests (ROIs) within the phantom. Secondly, in the global optimisation step, the fibres are divided into a set of control connected through truncated cones along their trajectories, which are then optimised concerning a global cost function inspired by [2,4]. The cost function includes features such as constrained curvature, smooth transversal radii changes in fibres, spring force between control points, or non-overlapping fibres. The optimised parameters are the control points' position and the circles' radii from the truncated cones. Finally, the meshing-growth step transforms the tubular-shaped fibres into more complex shapes using a post-processing framework based on BFS growth[13]. Each fibre is built in a local-sparse grid and then meshed with marching cubes[15] algorithms. Moreover, CACTUS introduces a new extension in the substrate construction that can generalise to create and extend preexisting large configurations like the DiSCo dataset[12] to be optimised and meshed.Results and Discussion
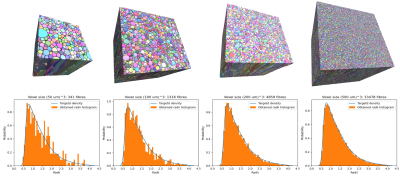
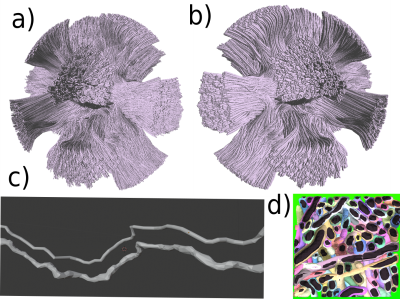
Single bundle substrates generated are shown in Figure 2, with cube substrates of length sizes 50, 100, 200, and 500 μm and an ICVF of 95%. The results show an increase in substrate sizes ranging from 35 to 100 μm per side and an ICVF from 65-70% [3, 4, 8]. The increased intracellular volume fraction and substrate size allow for an improved fit of the target distribution, chosen before the optimisation, as Figure 1 shows. Moreover, the new packing density percentage is comparable to previous histological studies [17,18,19], and the inter-axonal diameter of the synthetic fibres changes along the trajectories, as shown in 3D synchrotron segmentation studies[10].Figure 3 displays the differences between the cylindrical fibres used in DiSCo and those obtained with the CACTUS method. Our method adds extra complexity to the fibre meshing, allowing tortuous surface reconstructions and more dense packing. Figure 4 further shows the details of increased fibre packing, comparing the tubular shapes compared to the meshes optimised with the CACTUS method.
Conclusion
Numerical substrates have an increasingly higher role in microstructure modelling. The complexity of the white matter axons structure makes it challenging for state-of-the-art methods, particularly when it comes to constructing realistic and densely packed substrates. In this work, we provide a unique architecture for creating realistic and ultra-packed substrates. The improvement in density packing, fibre shape, and substrate sizes are features that will lead to improved realism of Monte-Carlo simulations in DW-MRI. Our initial findings show its capability of generating configurations with intra-axonal volume fractions as high as 95%. Furthermore, our substrates have microstructural properties that are essential for modelling the white matter microstructure [9]. Future research will expand the white matter substrate generation to include additional cell compartments, such as astrocytes and glial cells.Acknowledgements
This work is partly supported by the Swiss National Science Foundation under grant Nbr 205320_175974. We acknowledge access to the facilities and expertise of the CIBM Center for Biomedical Imaging, a Swiss research center of excellence founded and supported by Lausanne University Hospital (CHUV), University of Lausanne (UNIL), Ecole Polytechnique Fédérale de Lausanne (EPFL), University of Geneva (UNIGE), Geneva University Hospitals (HUG), and Université de Sherbrooke. Erick J. Canales-Rodríguez was supported by the Swiss National Science Foundation (SNSF, Ambizione grant PZ00P2_185814).References
[1] Blinn, James F. "A generalization of algebraic surface drawing." ACM transactions on graphics (TOG) 1.3 (1982): 235-256.
[2] Close, Thomas G., et al. "A software tool to generate simulated white matter structures for the assessment of fibre-tracking algorithms." NeuroImage 47.4 (2009): 1288-1300.
[3] Callaghan, Ross, et al. "ConFiG: Contextual Fibre Growth to generate realistic axonal packing for diffusion MRI simulation." Neuroimage 220 (2020): 117107.
[4] Ginsburger, Kévin, et al. "MEDUSA: A GPU-based tool to create realistic phantoms of the brain microstructure using tiny spheres." NeuroImage 193 (2019): 10-24.
[5] Andersson, M., Kjer, H. M., Rafael-Patino, J., Pacureanu, A., Pakkenberg, B., Thiran, J. P., ... & Dyrby, T. B. (2020). Axon morphology is modulated by the local environment and impacts the noninvasive investigation of its structure–function relationship. Proceedings of the National Academy of Sciences, 117(52), 33649-33659.
[6] Lee, Hong-Hsi, Els Fieremans, and Dmitry S. Novikov. "Realistic Microstructure Simulator (RMS): Monte Carlo simulations of diffusion in three-dimensional cell segmentations of microscopy images." Journal of Neuroscience Methods 350 (2021): 109018.
[7]Rafael-Patino, Jonathan, et al. "Robust Monte-Carlo simulations in diffusion-MRI: Effect of the substrate complexity and parameter choice on the reproducibility of results." Frontiers in neuroinformatics 14 (2020): 8.
[8] Rafael-Patino, J., Girard, G., Truffet, R., Pizzolato, M., Thiran, J. P., & Caruyer, E. (2021, October). The Microstructural Features of the Diffusion-Simulated Connectivity (DiSCo) Dataset. In International Workshop on Computational Diffusion MRI (pp. 159-170). Springer, Cham.
[9] Novikov, Dmitry S., et al. "Quantifying brain microstructure with diffusion MRI: Theory and parameter estimation." NMR in Biomedicine 32.4 (2019): e3998.
[10] Liewald, Daniel, et al. "Distribution of axon diameters in cortical white matter: an electron-microscopic study on three human brains and a macaque." Biological cybernetics 108.5 (2014): 541-557.
[11]Haro, Juan Luis Villarreal, et al. "Towards a high-density packing white matter substrate generator for Monte-Carlo simulations."
[12]Rafael-Patino, Jonathan, et al. "The Microstructural Features of the Diffusion-Simulated Connectivity (DiSCo) Dataset." International Workshop on Computational Diffusion MRI. Springer, Cham, 2021.
[13]Zhang, Hui, et al. "NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain." Neuroimage 61.4 (2012): 1000-1016.
[14]Bundy, Alan, and Lincoln Wallen. "Breadth-first search." Catalogue of artificial intelligence tools. Springer, Berlin, Heidelberg, 1984. 13-13.
[15]Lorensen, William E., and Harvey E. Cline. "Marching cubes: A high resolution 3D surface construction algorithm." ACM siggraph computer graphics 21.4 (1987): 163-169.
[16]Andersson, Mariam, et al. "Axon morphology is modulated by the local environment and impacts the noninvasive investigation of its structure–function relationship." Proceedings of the National Academy of Sciences 117.52 (2020): 33649-33659.
[17]Stikov, Nikola, et al. "In vivo histology of the myelin g-ratio with magnetic resonance imaging." Neuroimage 118 (2015): 397-405.]
[18]Abdollahzadeh, Ali, et al. "DeepACSON automated segmentation of white matter in 3D electron microscopy." Communications biology 4.1 (2021): 1-14.
[19]Tagge, Ian J., et al. "Permanent tissue damage in multiple sclerosis lesions is associated with reduced pre-lesion myelin and axon volume fractions." Multiple Sclerosis Journal (2022): 13524585221110585.
Figures