3019
A novel atlas of the superficial white matter connectivity based on the whole HCP cohort - an inter-individual variability study.1BAOBAB, NeuroSpin, Université Paris-Saclay, CNRS, CEA, Gif-sur-Yvette, France, 2AP-HP, Epilepsy Unit, GH Pitié-Salpêtrière-Charles Foix, Paris, France, 3Sorbonne Université, Paris, France
Synopsis
Keywords: White Matter, Brain Connectivity
Superficial white matter (SWM) tracts exhibit a high cross-subject variability, which makes them difficult to study. We used the whole HCP cohort, which included 1065 subjects, to build a comprehensive atlas of the short-range cortico-cortical connections using an advanced diffusion analysis and tractography pipeline, followed by an individual and a group-level fiber clustering. Furthermore, we measured the percentage of occurrence of each cluster within the population. In addition to enabling the automatic labeling of any new subject’s SWM tracts, our atlas paves the way for a thorough cross-subject variability study of superficial connectivity.Introduction
Superficial white matter (SWM) tracts constitute the anatomical substrate of major brain functions but their anatomy remains poorly known, because of the technical difficulties of their dissection with Klinger's technique, and their high variability inbetween subjects. Over the last decade, diffusion MRI allowed the generation of the first SWM atlases1–3. However, they were performed on reduced set of subjects due to the heavy computational and memory burden of fiber clustering techniques. We therefore addressed these technological issues and present a novel atlas of the SWM bundles from the whole Human Connectome Project (HCP) cohort.Method
Cohort and imaging protocol - We exploited the whole HCP MRI dataset4, corresponding to acquisitions performed on a Connectome Skyra 3T MRI system on a cohort of 1065 healthy subjects aged from 22 to 35 years. It includes, for each subject, an anatomical T1-weighted acquisition using a 3D MPRAGE sequence (0.7mm isotropic spatial resolution, TR/TE=2400/2.14 ms), and series of diffusion-weighted MRI (dMRI) sequences, using a 2D spin-echo single-shot multiband EPI sequence (multi-band factor of 3, monopolar diffusion gradient pulses, 1.25 mm isotropic spatial resolution, TR/TE=5500/89.50 ms) over 3 shells at b=1000/2000/3000 s/mm2 along 90 diffusion sampling directions for each shell, plus 6 non-diffusion-weighted (b=0 s/mm²) reference images. The data were already preprocessed and corrected for eddy current and susceptibility artifacts.Individual data processing - Using the Ginkgo toolbox (available at https://framagit.org/cpoupon/gkg, BAOBAB, NeuroSpin), we designed an analysis pipeline to infer the structural connectivity of each individual from their multiple-shell dMRI dataset (figure 1). It consists of the following steps: registration of the subject’s brain to the MNI ICBM152 2009c non-linear asymmetric template space with the ANTS (Advanced Normalization Tools) toolbox5,6 using the T1-weighted acquisition; computation of the diffusion Orientation Distribution Functions (ODF) map with the analytical Q-ball model7 from the dMRI; whole-brain fiber tracking using a regularized deterministic algorithm8 (1 seed/voxel, aperture angle: 30°, fiber length range: 1.25–300 mm, integration step: 0.3 mm); intra-subject fiber clustering with a hierarchical algorithm1,9 which groups fibers according to their geometrical properties.
Group analysis - A cross-subject fiber clustering was performed on the 1065 subjects’ fiber clusters, using the HDBscan algorithm10 (with the following parameters tuned to maximize the number of clusters: normalization factor 6, neighbor count 5, minimum cluster size 10, minimum subject percentage 2.5 %) modified to keep track of the subjects that contributed to each fascicle cluster. The low minimum subject percentage threshold enabled us to keep the fiber bundles in common even among a small percentage of the population, and thus being able to assess the rare anatomical variations of the superficial white matter tracts.
Reconstruction of SWM bundles - The fascicle cluster map computed at the previous step was filtered to keep only the clusters with a mean length shorter than 80mm. We then applied to it a pairwise selection process using the Desikan-Killiany atlas11 in order to extract clusters connecting two cortical regions of the atlas. This selection process also enabled us to discard all non-cortico-cortical connections. Ultimately, resulting SWM bundles were manually curated to remove any remaining artifactual tracts.
Results
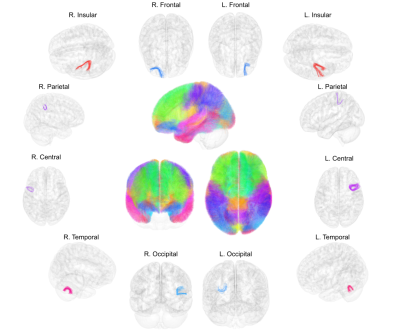
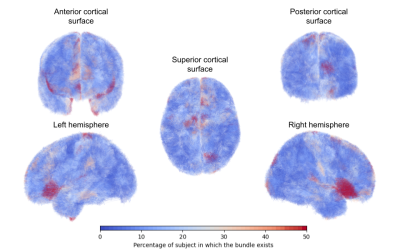
We reconstructed 638021 individual fascicles grouped into 3404 SWM bundles for the left hemisphere, and 632668 individual fascicles grouped into 3519 SWM bundles for the right hemisphere. The SWM atlas in the MNI space is displayed in figure 2, as well as 3D renderings of some typical individual SWM bundles. In order to further evaluate the anatomical cross-subject variability of SWM bundles at the surface of each hemisphere, we measured the percentage of occurrence of each cluster at the population level and colored the SWM bundles accordingly (blue color for the lowest representativity and red color for the highest representativity) (figure 3).Discussion
Superficial white matter bundles are hard to study due to their high inter-individual variability which may skew the results of cross-subject clustering. Indeed, cross-subject clustering, which seeks fibers in common among a population, is efficient to establish deep white-matter tract atlases because those tracts are highly conserved across individuals. On the other hand, superficial white matter tracts may largely differ from one individual to another, and the results of such clustering, as used in classical atlases12–16, only keep track of the most common ones in the population. New approaches suggest partitioning the population into subgroups according to their cortical folding patterns17 and performing a clustering on each subgroup. We preferred to perform a cross-subject clustering on a large population using a low frequency threshold (2.5 %), in order to keep the low frequency anatomical configuration of the superficial white matter, which results in more artifactual tracts to manually curate afterwards, but relies on fewer a priori assumptions.Conclusion
We established a novel superficial white matter atlas on a large cohort of 1065 subjects, with a new approach enabling to study cross-subject variability and anatomical variation of those tracts, including those found at a low frequency among the population. In the future, because of the outstanding number of subjects involved to establish our SWM atlas, we should be able to design new types of SWM bundle atlases relying on a stratification of the population, therefore better adapted to the segmentation of individual superficial brain connectivity of human subjects.Acknowledgements
Data collection and sharing for this project was provided by the Human Connectome Project (HCP; Principal Investigators: Bruce Rosen, M.D., Ph.D., Arthur W. Toga, Ph.D., Van J. Weeden, MD). HCP funding was provided by the National Institute of Dental and Craniofacial Research (NIDCR), the National Institute of Mental Health (NIMH), and the National Institute of Neurological Disorders and Stroke (NINDS). HCP data are disseminated by the Laboratory of Neuro Imaging at the University of Southern California.
Thanks to Ursi Zaiser for her help.
References
1. Guevara, P. et al. Automatic fiber bundle segmentation in massive tractography datasets using a multi-subject bundle atlas. Neuroimage vol. 61 1083–99 (2012).2. Guevara, M. et al. Reproducibility of superficial white matter tracts using diffusion-weighted imaging tractography. Neuroimage vol. 147 703–725 (2017).
3. Labra, N. et al. Fast Automatic Segmentation of White Matter Streamlines Based on a Multi-Subject Bundle Atlas. Neuroinformatics 15, 71–86 (2017).
4. Van Essen, D. C. et al. The WU-Minn Human Connectome Project: an overview. NeuroImage 80, 62–79 (2013).
5. Avants, B. B., Epstein, C. L., Grossman, M. & Gee, J. C. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal. 12, 26–41 (2008).
6. Avants, B. B. et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage 54, 2033–2044 (2011).
7. Descoteaux, M., Angelino, E., Fitzgibbons, S. & Deriche, R. Regularized, fast, and robust analytical Q-ball imaging. Magn. Reson. Med. 58, 497–510 (2007).
8. Perrin, M. et al. Fiber tracking in q-ball fields using regularized particle trajectories. Inf. Process. Med. Imaging Proc. Conf. 19, 52–63 (2005).
9. Guevara, P. et al. Inference of a HARDI fiber bundle atlas using a two-level clustering strategy. Med. Image Comput. Comput.-Assist. Interv. MICCAI Int. Conf. Med. Image Comput. Comput.-Assist. Interv. 13, 550–7 (2010).
10. Campello, R. J. G. B., Moulavi, D. & Sander, J. Density-Based Clustering Based on Hierarchical Density Estimates. in Advances in Knowledge Discovery and Data Mining (eds. Pei, J., Tseng, V. S., Cao, L., Motoda, H. & Xu, G.) 160–172 (Springer, 2013). doi:10.1007/978-3-642-37456-2_14.
11. Desikan, R. S. et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage 31, 968–980 (2006).
12. Oishi, K. et al. Human brain white matter atlas: identification and assignment of common anatomical structures in superficial white matter. NeuroImage 43, 447–457 (2008).
13. Román, C. et al. Clustering of Whole-Brain White Matter Short Association Bundles Using HARDI Data. Front. Neuroinformatics 11, 73 (2017).
14. Zhang, F. et al. An anatomically curated fiber clustering white matter atlas for consistent white matter tract parcellation across the lifespan. NeuroImage 179, 429–447 (2018).
15. Guevara, M. et al. Creation of a whole brain short association bundle atlas using a hybrid approach. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. IEEE Eng. Med. Biol. Soc. Annu. Int. Conf. 2016, 1115–1119 (2016).
16. Román, C. et al. Superficial white matter bundle atlas based on hierarchical fiber clustering over probabilistic tractography data. NeuroImage 262, 119550 (2022).
17. Guevara, M. et al. Disentangling the variability of the superficial white matter organization using regional-tractogram-based population stratification. NeuroImage 255, 119197 (2022).
Figures

Figure 1. Diffusion analysis pipeline.
Step 1: Individual diffusion analysis pipeline performed for each subject: raw diffusion data ➔ Q-Ball Imaging (QBI) and computation of RGB map and ODF field (color-coded direction: red=left-right; green=antero-posterior; blue, superior=inferior) ➔ whole-brain tractography ➔ intra-subject fiber clustering.
Step 2: Cross-subject fiber clustering using HDBScan algorithm.
Step 3: Parcellation of the superficial white matter according to cortical areas from Desikan-Killiany atlas