2997
Memorizing Transformer for Small-scale Multi-parametric MRI Brain Tumor Diagnosis
Yiqing Shen1,2, Nhat Le1,2, Jingpu Wu1,3, Pengfei Guo1,2, Jinyuan Zhou1, Mathias Unberath2, and Shanshan Jiang1
1Department of Radiology, School of Medicine, Johns Hopkins University, Baltimore, MD, United States, 2Department of Computer Science, Whiting School of Engineering, Johns Hopkins University, Baltimore, MD, United States, 3Department of Applied Mathematics and Statistics, Whiting School of Engineering, Johns Hopkins University, Baltimore, MD, United States
1Department of Radiology, School of Medicine, Johns Hopkins University, Baltimore, MD, United States, 2Department of Computer Science, Whiting School of Engineering, Johns Hopkins University, Baltimore, MD, United States, 3Department of Applied Mathematics and Statistics, Whiting School of Engineering, Johns Hopkins University, Baltimore, MD, United States
Synopsis
Keywords: CEST & MT, Machine Learning/Artificial Intelligence
Deep learning approaches have been widely applied to the MRI field. Among them, transformers have received increasing popularity due to their capability in handling multi modalities. Yet, transformers are hungry for large-scale data, which is expensive to collect. Here, we develop a novel memorizing transformer for small-scale multi-parametric MRI analysis. Empirically, we evaluate the proposed method on a dataset curated from 147 brain post-treatment malignant glioma cases for classifying treatment effect and tumor recurrence. The proposed memorizing transformer boosted a 5.15% improvement in the test area under the receiver-operating-characteristic curve (AUC) to the baseline transformer approaches.Introduction
Gliomas are the most common and lethal primary brain tumors in adults. Despite advances in therapy, malignant gliomas remain almost uniformly fatal. Reliably distinguishing tumor from treatment effect is of significant importance to patients and clinicians. It allows clinicians to determine the efficacy of the implemented treatment plan and enables real-time modifications to the regimen. Deep learning methods demonstrate their promising potential in a wide range scope of MR imaging research1. Among them, the transformer has gained the most attention from researchers due to its high capability in fusing multi-modalities, e.g., multi-parametric MRI sequences2. However, transformer is data-hungry, particularly when training with multi-modality data. Thus, its application has been hindered because acquiring large-scale multimodal MR data is expensive and time-consuming. In this work, we design a novel memorizing transformer model for the small-scale MR classification in neuro-oncology.Method
A dataset from 147 scans with malignant gliomas (dataset details in Table 1), each of which included five MRI sequences, T1-weighted (T1w), T2-weighted (T2w), fluid-attenuated inversion recovery (FLAIR), gadolinium-enhanced T1w (GdT1w), and amide proton transfer-weighted (APTw) MR images, was curated for this work. These MR images were acquired with the following parameters: i) APTw: RF saturation duration, 830 ms; saturation power, 2 µT; FOV, 212 × 186 × 66 mm3; resolution, 0.82 × 0.82 × 4.4 mm3. ii) T2w: TR, 4 sec; TE, 80 ms; 60 slices. iii) FLAIR: TR, 11 sec; TE, 120 ms; inversion recovery time, 2.8 sec; 60 slices. iv) T1w and GdT1w: 3D MPRAGE; TR, 3 sec; TE, 3.7 ms; inversion recovery time, 843 ms; 150 slices. The anatomic MRI sequences (T1w, T2w, FLAIR, and GdT1w) had FOV, 212 × 192 × 132/165 mm3; resolution, 0.41 × 0.41 × 2.2/1.1 mm3. For each scan, the 3D APTw MRI protocol provided 15 slices, so all volumetric MR images used 15 instances. Each instance included T1w, T2w, FLAIR, GdT1w, and APTw images with the matrix shape of 5 (sequences) × 256 (pixels) × 256 (pixels). Instances were the input of the proposed slice-level transformer.A proportion of 80% on the case-level was first split as the training set (n=118) and the remaining 20% as the test set (Table 1). We used a fixed number of training epochs; thus, there is no need for a validation split. Both our approach and baseline were trained and evaluated at the slice-level. We employed the area under the receiver-operating-characteristic curve (AUC), sensitivity, specificity, and accuracy as the evaluation metric by receiver operating characteristic curve (ROC) analysis, and 95% confidence intervals (CI) using a bootstrap were recorded.
To close the gap for the data-hungry nature of the transformer, we designed a transformer architecture with external memory, which memorizes the attention from the historical training process4. To be more specific, the external memory stores a paired query and value from each attention head during the training (overall framework in Fig. 1). It follows that this design will not add to extra computation cost during the training. In the inference, extra attention will be retrieved from the cached memory by K nearest neighbor (KNN) lookup to enhance the local attention. We implement the external memory on vision transformer (ViT)5, but this design with external memory can be plugged into any transformer-based model.
Results
We implemented the method on an 8-layer ViT, with 8 attention heads in each layer and the dimension of the tokens were set to 256. We set K = 32 in KNN lookup. The baseline was the ViT without external memory. The corresponding AUC, sensitivity, specificity, and accuracy were listed in Table 2. Our method achieved an AUC of 0.8484 (p < 0.001), which improves 5.15% of the plain ViT’s AUC of 0.8068 (Fig. 2, p < 0.001).Conclusion
In this work, we propose a memorizing transformer for small-scale multi-parametric MR image analysis. The method stores the attention in the training process and applies them to the inference stage with KNN lookup, which thus costs no extra GPU run-in memory. Empirically, the method can boost the plain transformer by 5.15%, which confirms the effectiveness of our method.Acknowledgements
The authors thank our clinical collaborators for help with the patient recruitment and MRI technicians for assistance with MRI scanning. This study was supported in part by grants from the NIH.References
- Lee, et al. "Deep learning in MR image processing." Invest Magn Reson Imaging 23:81-99(2019).
- He, et al. "Transformers in medical image analysis: A review." arXiv:2202.12165 (2022).
- Guo, et al. "Learning-based analysis of amide proton transfer-weighted MRI to identify true progression in glioma patients." NeuroImage: Clinical 35:103121 (2022).
- Wu, et al. "Memorizing transformers." arXiv:2203.08913 (2022).
- Dosovitskiy, et al. "An image is worth 16x16 words: Transformers for image recognition at scale." arXiv:2010.11929 (2020).
Figures
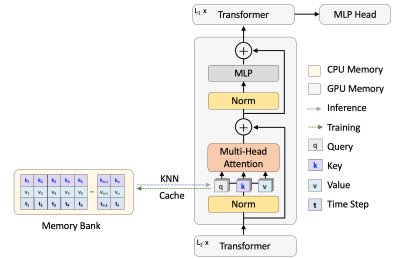
Fig. 1 The overall framework for the memorizing transformer.
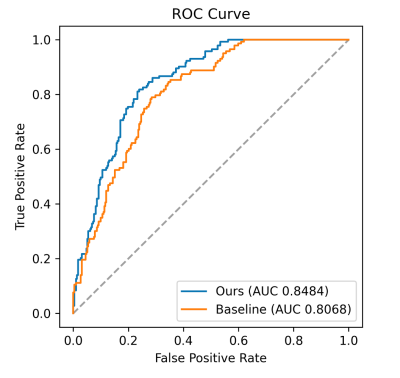
Fig. 2 ROC comparison between memorizing transformer with plain ViT.
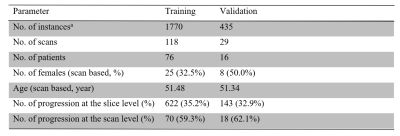
Table.1 Participant Demographic Information, and Basic Characteristics of the Datasets.
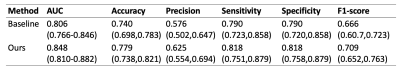
Table.2 The diagnostic performances of the baseline transformer (ViT) and our proposed approach.
DOI: https://doi.org/10.58530/2023/2997