2984
Highly Accelerated Chemical Exchange Saturation Transfer Imaging with Partially Separable Network1Center for Biomedical Imaging Research, Department of Biomedical Engineering, Tsinghua University, Beijing, China, 2Institute of Science and Technology for Brain-Inspired Intelligence, Fudan University, Shanghai, China
Synopsis
Keywords: CEST & MT, Machine Learning/Artificial Intelligence
Herein, we developed a partially separable network (PSN) for CEST acceleration. Our contributions are: 1) We found that the reconstruction error of CEST mainly exists in the spatial subspace. 2) A deep learning network based on partially separable model was developed to optimize CEST images in spatial subspace. Retrospective results suggested that our method enabled a highly accelerated CEST imaging (14X for healthy adults and 11X for brain tumor patients) with contrast maps and Z-spectrum consistent with gold standard, which could have great clinical utility.Introduction
Chemical Exchange Saturation Transfer MRI is a promising ‘label-free’ molecular imaging tool available in clinical applications. Usually CEST requires collection of several saturation weighted images, each with a seconds-long saturation preparation for signal amplification. Therefore, current acquisition protocol faces obstacles of long scan time, low-resolution images and limited number of saturation frequencies[1][2].In this study, we developed a partially separable network (PSN) for CEST acceleration. Our contributions are: 1) We found that the reconstruction error of CEST mainly exists in the spatial subspace. 2) A deep learning network based on partially separable model was developed to optimize CEST images in spatial subspace. Retrospective results on healthy volunteers and patients indicated that PSN enables a highly accelerated CEST imaging, while ensuring the accuracy of contrast maps obtained from widely used quantification methods.
Methods
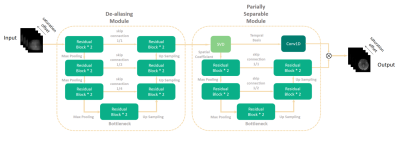
Principle of Partially Separable NetworkHere we introduce the principle of partially separable[3][4] network. The network consists of two modules (Figure 1). The first module is a U-net based structure, which takes linear reconstructed multi-contrast saturated images as input and dealiasing them initially. The network are 3-dimensional to take advantages of 3D operators’ capacity on local pixel-wise correlation learning along spatial and frequency dimensions. Despite the de-aliasing module initially reconstructed de-aliased images, it is not capable of producing accurate contrast maps which are sensitive to noise. Thus, we developed a partially separable module to further optimize images in spatial and temporal subspace. Since Z-spectrum of CEST images have very high similarity, reconstructed image from de-aliasing module can be further expressed in decomposed subspace as:
$$C_{1}\left(X_{\text {aliased }}\right)=U \Sigma V^{T}=ST=\left(\widehat{S}+n_{1}\right)\left(\widehat{T}+n_{2}\right)\approx\left(\widehat{S}+n_{1}\right) \widehat{T}$$
$$\text{where}S=U\Sigma^{0.5},T=\Sigma^{0.5}V^{T}$$
where C1 denotes the de-aliasing mapping, U and V denote spatial coefficient and temporal basis respectively, n1 and n2 are remaining noise in spatial and temporal subspace. For well-de-aliased images with little noise, temporal basis obtained from singular value decomposition are almost noise-free because of the large sample with thousands of pixel-wised Z-spectra. Therefore, we assume that for CEST acceleration, data in spatial subspace determines the accuracy of contrast maps. The forward model can be expressed as:
$$X=C_{2w}(S)C_{2s}(T)$$
where C2w is a one-dimensional convolutional network for optimizing the temporal basis while C2s is a 3-layer 3D Res-Unet that learns spatial coefficients on the spatial subspace and determines the component weights of all temporal bases. Note that the initial S and T were SVD decomposition of C1 output. The outputs of two block were multiplied after separate optimization. Above this, the optimization objective is calculated as:
$$\text{loss}=\min_{\theta_{1},\theta_{2 w},\theta_{2s}}\left\{\lambda\left\|\widehat{X}-C_{2 w}\left(\theta_{2 w}\mid S\right)C_{2 s}\left(\theta_{2 s}\mid T\right)\right\|_{2}+(1-\lambda)\left\|\widehat{X}-C_{1w}\left(\theta_{1}\mid X_{\text {alised }}\right)\right\|_{2}\right\}$$
Data Acquisition & Preprocessing
13 healthy volunteers and 15 brain tumor patients were scanned on 3.0 Tesla scanner (Ingenia, Philips Healthcare) with a 32-channel phase array coil. All volunteers signed the written informed consent. For each MRI study, a 3D TSE CEST sequence was executed 30 times with saturation applied at frequencies of -10 to 10 ppm relative to water. Other scan parameters are: FOV=230×201×60 mm3, spatial resolution=1.8×1.8×3.3 mm3, TR/TE=3.39 s/7.8 ms.
In terms of data preprocessing, gold standard labels were linearly reconstructed from fully sampled k-space data. For retrospective research, we undersampled k-space data with variable density catesian sampling pattern. Training and testing data were normalized before training and finally we got a CEST dataset with 243 paired 3D cases. Dataset was randomly split in a ratio of about 9:1:2.
Network Training
In terms of network training, the optimizer was Adam and the learning rate was set to 1e-3, then decayed by 10 times at 30 and 150 epoch. We employed the pixel-wise mean squared error (MSE) as loss function. The two modules reached convergence after 300 epochs without overfitting.
Results and Discussion
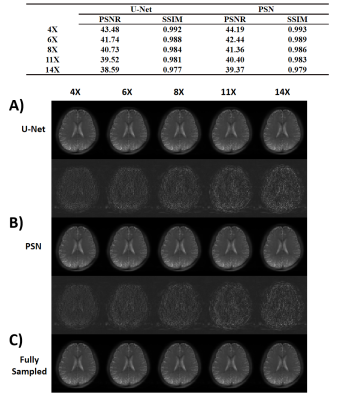
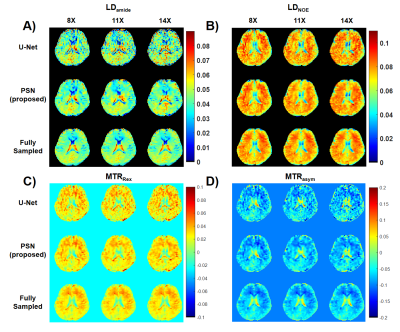
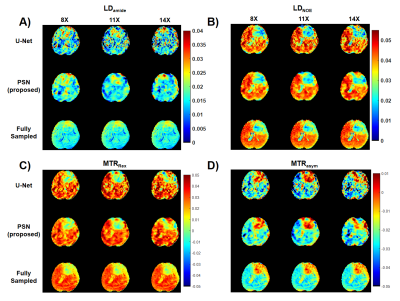
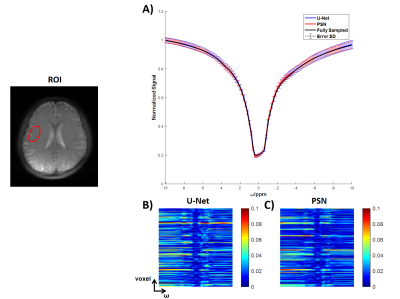
We evaluated the performance of PSN, with regular Fourier-transform based reconstruction from fully-sampled data as the golden standard. Figure 2 compared the saturation-weighted images at 3.5 ppm obtained from the full-sampled data, the PSN reconstruction and U-Net[5] reconstruction from the retrospective data, respectively. The error between PSN reconstructed images and fully-sampled golden standard was lower at all acceleration ratios, which suggesting the good recovery of anatomy structure. We further calculated the contrast maps of healthy adults and tumor patients through the widely-used quantification methods, i.e., MTR asymmetry, Lorentzian difference (LD) and MTR Rex. As shown in Figure 3-4, U-Net reconstruction were noisy and displayed altered contrast from the golden standard while the proposed PSN suggested very closed contrast maps at high acceleration ratio (14X for healthy adults and 11X for tumor patients).Figure 5 depicted the averaged Z-spectrum in a brain region ROI and corresponding error maps at -10 to 10 ppm. The proposed PSN can produce Z-spectra consistent with the gold standard and with better accuracy than U-Net. Note that the U-Net's Z-spectrum were also closed to gold standard. However, its contrast maps were noisy, which illustrated the advantage of the proposed subspace optimization.
Conclusion
In this study, we developed a deep learning based partially separable reconstruction network for CEST acceleration at 3T human scanners. Retrospective results suggested that our method enabled a highly accelerated CEST imaging (14X for healthy adults and 11X for brain tumor patients) with contrast maps and Z-spectrum consistent with gold standard, which could have great clinical utility.Acknowledgements
This work was supported by the National Natural Science Foundation of China [grant numbers 82071914]References
[1] Y. Zhang, H.-Y. Heo, D.-H. Lee, S. Jiang, X. Zhao, P. A. Bottomley, et al., "Chemical Exchange Saturation Transfer (CEST) Imaging With Fast Variably-accelerated Sensitivity Encoding (vSENSE)," Magnetic Resonance in Medicine, vol. 77, pp. 2225-2238, Jun 2017.
[2] C. Guo, J. Wu, J. T. Rosenberg, T. Roussel, S. Cai, and C. Cai, "Fast chemical exchange saturation transfer imaging based on PROPELLER acquisition and deep neural network reconstruction," Magnetic Resonance in Medicine, vol. 84, pp. 3192-3205, Dec 2020.
[3] H. She, J. S. Greer, S. Zhang, B. Li, J. Keupp, A. J. Madhuranthakam, et al., "Accelerating chemical exchange saturation transfer MRI with parallel blind compressed sensing," Magnetic Resonance in Medicine, vol. 81, pp. 504-513, Jan 2019.
[4] M. Lustig, D. Donoho, and J. M. Pauly, "Sparse MRI: The application of compressed sensing for rapid MR imaging," Magnetic Resonance in Medicine, vol. 58, pp. 1182-1195, Dec 2007.
[5] X. Xiao, S. Lian, Z. Luo, S. Li, and Ieee, "Weighted Res-UNet for High-quality Retina Vessel Segmentation," in 9th International Conference on Information Technology in Medicine and Education (ITME), Hangzhou, PEOPLES R CHINA, 2018, pp. 327-331.[6] Zhao B, Haldar JP, Liang ZP. PSF model-based reconstruction with sparsity constraint: algorithm and application to real-time cardiac MRI. Annu Int Conf IEEE Eng Med Biol Soc. 2010;2010:3390-3. doi: 10.1109/IEMBS.2010.5627934. PMID: 21097243; PMCID: PMC3121182.
Figures