2953
Deep learning-based saturation band placement in spine localizer images
Ashish Saxena1, Chitresh Bhushan2, Soumya Ghose2, Patil Uday1, Kameswari Padmanabhan3, Sanjay NT3, and Dattesh Shanbhag3
1GE Research, Bengaluru, India, 2GE Research, Niskayuna, NY, United States, 3GE Healthcare, Bengaluru, India
1GE Research, Bengaluru, India, 2GE Research, Niskayuna, NY, United States, 3GE Healthcare, Bengaluru, India
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Visualization, workflow
Saturation band placement is important for obtaining good quality MRI images in presence of structures which can generate artifacts such as cardiac regions or large pulsating blood vessels. In anatomy such as spine, saturation-band placement can be time consuming since technologist has to ensure that it doesn’t overlap with vertebrae regions. In this study, we demonstrated an automated deep-learning method to accurately place the saturation band on 2D three-plane localizer images with no intervention from the technologist.Introduction
In MRI, saturation bands (SB) are spatially selected and applied to suppress signal from unwanted regions. They help in reducing motion artifacts by avoid regions of flow or motion in the imaging slice. This requires additional setup time and skills for accurate SB placement [1]. In the literature, automation of MRI scan plane prescription has been demonstrated [2-3], which allows for contiguous visualization of anatomical landmarks, irrespective of changes in patient pose, minor anatomical changes, and technologist training. However, no such automation is yet attempted in case of SB placement. In this study, we trained a deep learning model to accurately place the saturation band in the spine localizer images.Methods
Subjects: Spine MRI data was collected from multiple sites. A total of 123 cervical and 181 lumbar scans from volunteers as well as patients were included in the study. All the studies were approved by respective IRBs.MRI Scanner and Acquisition: Three-plane 2D localizer Data was acquired on multiple MRI scanners (GE 3T Discovery MR 750w, GE Signa HDxt 1.5T, GE 1.5T Optima 450w) and with different coil configurations. Only single-shot fast spin echo (SSFSE) based localizer data were considered but had variations in imaging FOV and coverage, image resolution and matrix size across subjects.
Ground Truth (GT) labelling: Since localizer images are low-resolution, we first made use of the high-resolution sagittal T1W images to segment the spine vertebrae and intra-vertebra disc (IVD) using our in-house deep learning-based tool. Thereafter, using rigid image registration (Elastix) [3], localizer images were aligned to high resolution images and the spine vertebrae and IVD segmentations transferred to localizer images. These segmentations were then used as a reference to place a SB plane mask (Figure 1).
Deep learning (DL) model training: For each of the three spine stations, namely, cervical, lumbar, and sacral, a separate saturation band placement model was trained. We leverage a similar framework as suggested in [4] for segmenting the saturation band plane mask (3D U-Net model, dice loss function). The dataset used for the training was augmented five times using random rotations, cropping and voxel intensity changes.
Model Evaluation: Since this is a semantic segmentation learning, Dice score was used. It should be noted that even though the model was trained using a dice loss, it is not the right accuracy metric for the entire SB placement process, as the DL segmentation will be used to generate the saturation band. Therefore, in addition to dice score, three metrics were used to evaluate the performance of the model. First, mean absolute distance (MAD) error was calculated between GT and DL-estimated planes, which quantifies mean distance between two planes. Second, the center error between GT and DL-estimated planes was reported. Third, the minimum distance (MD_V) between DL-estimated planes and spine vertebrae was calculated. MAD error and center error < 3mm and MD_V > 0 were considered as acceptable for SB placement.
Results and Discussion
Our DL model learns the spine anatomical structure for SB placement. Figure 2 shows the results of model predictions at cervical, lumbar, and sacral stations. The predicted voxels will be used to fit a plane that will give the DL-estimated plane for SB placement in the localizer images. Although the mean dice score of these segmentations is low (Figure 3a), other relevant metrics strongly support the high accuracy of the model predictions. The MAD error and center for DL model prediction at all the spine stations were found to be lower than 3 mm (Figure 3b). There are few cases with outliers or error value of more than 3mm observed. However, given the value of MD_V was found to be greater than 0 in all the cases, it can be established that the SB plane predicted by the DL model doesn’t intersect with the respective spine station vertebrae (Figure 3c).Conclusions
We demonstrated a DL-based saturation band placement, in MRI localizer images, at cervical, lumbar, and sacral stations of spine. The results show an accurate placement of the saturation band using our DL model. The model performance is evaluated using quantitative parameters pertaining to the definition of saturation band in the spine. This can be easily adapted in the clinical practice that will result in reducing the scan time and technologist training efforts.Acknowledgements
NAReferences
- Serai, Suraj D., et al. "Newly developed methods for reducing motion artifacts in pediatric abdominal MRI: tips and pearls." American Journal of Roentgenology 214.5 (2020): 1042-1053.
- Lecouvet FE, Claus J, Schmitz P, Denolin V, Bos C, Vande Berg BC. Clinical evaluation of automated scan prescription of knee MR images. Journal of Magnetic Resonance Imaging: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2009 Jan;29(1):141-5.
- S. Klein, M. Staring, K. Murphy, M.A. Viergever, J.P.W. Pluim, "elastix: a toolbox for intensity based medical image registration," IEEE Transactions on Medical Imaging, vol. 29, no. 1, pp. 196 - 205, January 2010.
- Shanbhag DD et.al. A generalized deep learning framework for multi-landmark intelligent slice placement using standard tri-planar 2D localizers. In Proceedings of ISMRM 2019, Montreal, Canada, p. 670.
Figures
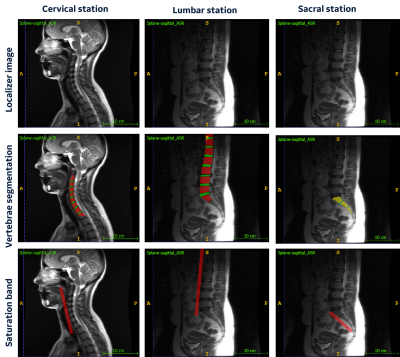
Figure
1: Localizer images from cervical, lumbar, and sacral
station of the spine with localizer registered vertebrae and intra-vertebra
disc (IVD) segmentation from corresponding high-resolution images
and placement of ground truth saturation band in each of the stations.
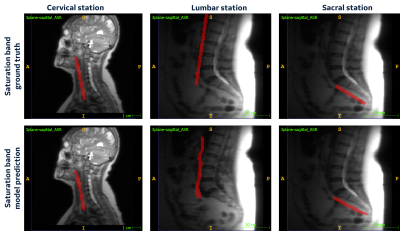
Figure
2: Cervical, Lumbar and Sacral localizer images with
ground truth saturation band and model predicted saturation band voxels.
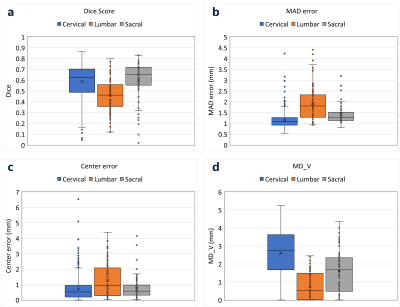
Figure 3: Box plot for model evaluation metrics a) Dice score:
pixel level match between model prediction and ground truth labels b) MAD error
(mm): mean distance between ground truth and
model-estimated planes c) Center error (mm): center point distance between
ground truth and model-estimated planes d) MD_V (mm): minimum distance
between model-estimated planes and spine vertebrae
DOI: https://doi.org/10.58530/2023/2953