2952
Deep Learning based prediction of the planes for automated planning of MRI imaging of cervical neural foramina and lumbar pars interarticularis
Chitresh Bhushan1, Dattesh D Shanbhag2, Uday Patil2, Trevor Kolupar3, and Maggie Fung3
1AI and Medical Imaging, GE Research, Niskayuna, NY, United States, 2GE Healthcare, Bangalore, India, 3GE Healthcare, Waukesha, WI, United States
1AI and Medical Imaging, GE Research, Niskayuna, NY, United States, 2GE Healthcare, Bangalore, India, 3GE Healthcare, Waukesha, WI, United States
Synopsis
Keywords: Machine Learning/Artificial Intelligence, Visualization, Scan Planning, Spine, Reformatting
We present a generalized DL-based intelligent slice placement framework for planes of cervical neural foramina (CF) and lumbar pars interarticularis (PI) for spine MRI. CF and PI scan improves assessment of foraminal stenosis and lumbar spondylolysis respectively, but requires highly skilled operator for accurate prescription. Our approach enables automatic patient-specific scan plane prescription from routine axial T2W images. In our test, it achieves mean error of <0.7 mm and <0.2 degrees and demonstrates similar/better contiguous anatomical visualization to manual scans on retrospective reformatting of 3D data. These results indicate that our approach is accurate and suitable for clinical usage.Introduction
Deep learning (DL) based intelligent scan plane (ISP) prescription has demonstrated good repeatability and improved performance vis-à-vis manual prescription for various landmarks across anatomies1-4. In this work, we have leveraged a similar approach for automating the prescription of oblique planes across the long axis of cervical neural foramina (CF) and oblique plane along lumbar pars interarticularis (PI). The oblique CF scan plane perpendicular to the long axis of foramen provides best assessment of cross-sectional area reducing inter-observer variabilities for assessment of foraminal stenosis compared to the standard sagittal or axial images of the spine5,6. Similarly, oblique PI plane improves assessment of pars interarticularis defects in lumbar spine as compared to the sagittal and axial planes alone7. Automated prescription of these scan-planes would be very impactful to reduce variability in acquisition and setup time, irrespective of technologist familiarity with spine anatomy. Compared to previous DL-based ISP methods1-3, we have utilized regular T2w axial images, instead of localizers.Methods
Data acquisition: 454 cervical and lumbar spine exams (IRB approved) from two clinical sites were used, which included axial 2D T2w spine images (multi-slice multi-angle (MSMA) or block acquisitions) over multiple vertebrae. Data came from various GE 1.5T and 3.0T MRI scanners with varying protocol parameters. Additionally in two volunteers, a trained technologist acquired 3D sagittal left and right cervical foramina data (3D CUBE, TR:2000, TE: 90.7, 210x210x40mm3 FOV, 0.2x0.20.5mm3 resolution) as well as a 3D axial data stack (3D CUBE, TE: 90.3, TR: 3479, 180x180x80mm3 FOV, 0.35x0.35x1mm3 resolution). The plane prediction was done on 3D axial stack and was reformatted accordingly using the predicted plane.Ground-truth (GT) marking: A trained radiologist marked the plane of CF and PI on both the left and right side (with different labels) of the landmark planes (Fig.1).
DL Data Preparation: To account for variations in slice thickness and angulations from MSMA stack, the GT marking was extended to neighboring slices using 1D dilation along slice direction (Fig.1B). The data was then cropped to retain only the center region of the axial image. All data was resampled to 256x256 matrix size and several intensity-augmentation were applied, including coil sensitivity, smoothening and sharpening filters etc. SimpleITK8 was used for data preparation and post-processing.
DL Methodology: For CF, a total of 116 Train (1326 augmented volumes), 18 Validation (208 augmented volumes) and 19 test subjects were available. For PI, a total of 200 Train (818 augmented volumes), 28 validation (117 augmented volumes) and 85 test cases were available. For segmentation of CF and PI scan planes, a variant of U-Net architecture1, was adapted with four layers of dyadic reduction and expansion with skip connections. The loss function was a combination of dice coefficient and boundary distance loss9. The dice loss function was the primary loss function till the boundary loss was >0 and then combined with boundary loss using weight factors as: 0.33 dice, 0.67 : Boundary loss.
Accuracy Assessment: The trained model predicted binary mask for CF and PI scan planes which comprises both the left and right planes. These were separated using image center information to obtain separate left and right scan planes. Analytical form of the scan planes were obtained by fitting planes to the predicted scan plane mask. Accuracy was assessed by calculating mean absolute distance (MAD) error and angle error between GT and DL-predicted planes for all the landmarks. MAD error <1 mm and angle-error <3⁰ was considered as acceptable for ISP.
We also retrospectively reformatted the 3D axial CUBE data using the predicted CF scan plane for left and right directions to generate sagittal view and compared it to manually prescribed sagittal CF plane prescriptions. The results were reviewed with a radiologist to ascertain the clinical acceptance.
Results and Discussion
For DL based CF prediction model, the angle and MAD errors for both CF and PI planes along right and left direction were within the acceptable limits (Fig 2). Fig 3 shows the prediction of CF plane on the two volunteers datasets. The DL-predicted planes were used to fit analytical plane for reformatting the isotropic axial 3D data. Fig 4 and 5 demonstrate the results of reformat and compare to manually prescribed CF plane. We notice that in both the cases, the DL-based CF plane reformatted data is similar or slightly better in providing a contiguous visualization of foramina; especially with a straight spine. In case of curved spine (Fig 5), a single plane will not be able to fit the entire foramina region and this is evident in both the manual and DL-based plane prescription. However, in clinical practice, the foramina plane will be typically acquired around C4-C5 region (and similarly L4-L5 region for PI plane) and hence results are acceptable for clinical prescription.Conclusion
We adapted a generalized DL-based intelligent slice placement framework for automated planning of cervical foramina and pars interarticularis for MRI spine exams. It achieved mean error of <0.7 mm and <0.2 degrees in our test. Prospective reformatting of 3D data demonstrates similar/better contiguous visualization of anatomical structure as compared to manual scan. These results indicate that our framework allows for patient specific automated plane prescription, which can be used in clinical practice.Acknowledgements
No acknowledgement found.References
- Shanbhag et al. A generalized deep learning framework for multi-landmark intelligent slice placement using standard tri-planar 2D localizers. In Proceedings of ISMRM 2019, Montreal, Canada, p. 670.
- Bhushan et al., Intelligent Knee MRI slice placement by adapting a generalized deep learning framework, ISMRM & SMRT Virtual Conference & Exhibition, p. 3562, (2020)
- Bhushan et al., Consistency in human and machine-learning based scan-planes for clinical knee MRI planning, ISMRM & SMRT Virtual Conference & Exhibition, p. 0675, (2020)
- Lecouvet FE, Claus J, Schmitz P, Denolin V, Bos C, Vande Berg BC. Clinical evaluation of automated scan prescription of knee MR images. Journal of Magnetic Resonance Imaging: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2009 Jan;29(1):141-5.
- Park, Moon Soo, et al. "Diagnostic value of oblique magnetic resonance images for evaluating cervical foraminal stenosis." The Spine Journal 15.4 (2015): 607-611.
- Winegar BA, Kay MD, Taljanovic M. Magnetic resonance imaging of the spine. Pol J Radiol. 2020 Sep 25;85:e550-e574. doi: 10.5114/pjr.2020.998877. DW Johnson, GN Farnum, RE Latchaw, and SM Erba, American Journal of Roentgenology 1989 152:2, 327-332
- Standaert, C. J., and S. A. Herring. "Spondylolysis: a critical review." British journal of sports medicine 34.6 (2000): 415-422.
- Lowekamp, Bradley C., et al. "The design of SimpleITK." Frontiers in neuroinformatics 7 (2013): 45.
- Kervadec, Hoel, et al. "Boundary loss for highly unbalanced segmentation." International conference on medical imaging with deep learning. PMLR, 2019.
Figures
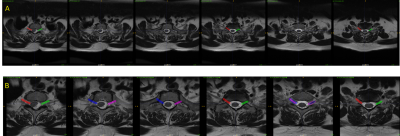
Figure 1: Panel A indicates the ground-truth marking by radiologist on lumbar Axial T2 images for PI plane (right and left side are marked differently). Panel B indicates the data prepared for deep learning-based segmentation (cropping and 1D dilation along slice direction).
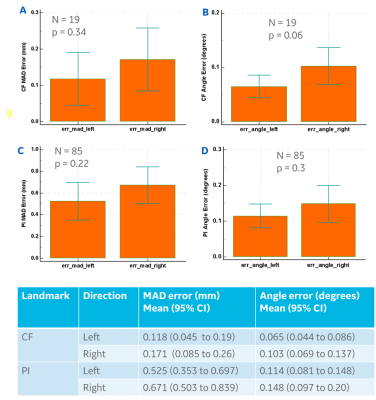
Figure 2: Summary of mean absolute distance (MAD) error and angle error for (Panel A, B) cervical neural foramina (CF) and (Panel C, D) lumbar pars interarticularis (PI) with respect to radiologist marked ground-truth planes along the right and left directions. Bottom panel shows the mean and 95% confidence interval (CI) for all accuracy metrics. There was no statistical difference between accuracy of predicted planes on left and right directions as computed by paired t-test (p-value included in plots).
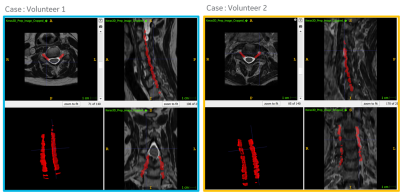
Figure 3: Example prediction of cervical neural foramina (CF) planes on cervical axial T2w data for two volunteers using the proposed approach. A 3D rendering of the predicted plane masks are also included in bottom left panel for reference.
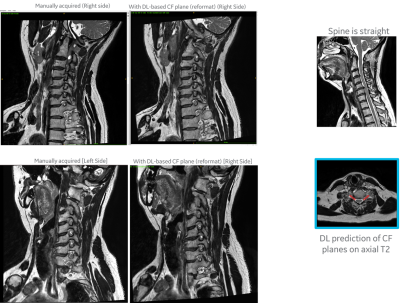
Figure 4: Volunteer #1 (straight spine) - Qualitative comparison of cervical neural foramina (CF) imaging as obtained with (left column) manual prescription, and (middle column) reformatted 3D data using our DL-predicted CF plane. Notice that both
are almost in-distinguishable, with DL predicted reformatted image having
better foramina along the inferior side and more vertically oriented spine. This subject has a straight spine
which enable good visualization of foramina along the length of the spine (right column).
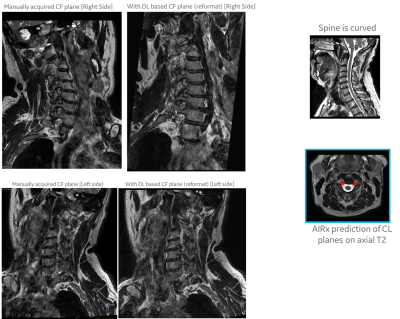
Figure 5: Volunteer #2 (curved spine): Qualitative comparison of cervical neural foramina (CF) imaging as obtained with (left column) manual prescription, and (middle column) reformatted 3D data using our DL-predicted CF plane. This subject has a curved spine and hence there is no single plane which will enable good visualization of foramina along the entire length of the spine. But manual and DL predicted prescriptions are almost same in this subject as well.
DOI: https://doi.org/10.58530/2023/2952