2946
Generative diffusion model for DSC MRI artifact correction
Muhammad Asaduddin1, Eung Yeop Kim2, and Sung-Hong Park1
1Bio and Brain Engineering, Korea Advanced Institute of Science and Technology, Daejeon, Korea, Republic of, 2Department of Radiology, Samsung Medical Center, Sungkyunkwan University College of Medicine, Seoul, Korea, Republic of
1Bio and Brain Engineering, Korea Advanced Institute of Science and Technology, Daejeon, Korea, Republic of, 2Department of Radiology, Samsung Medical Center, Sungkyunkwan University College of Medicine, Seoul, Korea, Republic of
Synopsis
Keywords: Data Processing, DSC & DCE Perfusion
Dynamic susceptibility contrast (DSC) MRI may suffer from artifacts due to long acquisition time. Past methods are limited in their performance and may change the contrast passage timing. In this work, we present a generative diffusion model that can restore signal loss and movement artifacts. We showed the generated DSC MRI images to have proper post-contrast vessel and grey matter structure with accurate contrast agent arrival/washout timing. The brain shape was also accurately generated as shown by the DICE score. This approach could provide a solution to restore a corrupted DSC MRI data while maintaining accurate contrast passage timing.Introduction
Dynamic susceptibility contrast (DSC) MRI scans often take 2 minutes or longer to acquire while requiring the patient to stay at the same position. Due to the long acquisition time, many artifacts including motion and signal loss can corrupt the DSC MRI data. Artifact correction for DSC MRI data is often limited and may fail to reflect the contrast agent-related enhancement (1,2). In this study, we propose a generative diffusion model-based approach to restore the corrupted scans in DSC MRI.Method
DSC MRI data from 60 patients with varying degree of abnormalities were used in this study, 50 of which are reserved for training and 10 for test. DSC MRI data were acquired using 2D EPI sequence with the following parameters: Acquisition matrix = 128x128x24, FOV = 230x230x134.4mm3, thickness = 4mm, number of dynamic scans = 50.Three different data corruptions were synthetically introduced to the DSC MRI data. Movement artifacts are introduced by rotating the DSC MRI image at random time points at the rotation angle ranging from -5 to 5 degree. Another data corruption was introduced at random time points in dynamic scans by replacing with noise. Using the two corruption methods, we generated three different DSC MRI corruption cases: Movement + Noised, Movement, and Noised DSC MRI. The corrupted DSC MRI data was generated for each training step, thus the corrupted time points were not repetitive and randomized.
A generative diffusion model was designed to generate a non-corrupted DSC MRI dynamic data using the corrupted DSC MRI dynamic data as the conditioning input. Denoising diffusion probabilistic model (DDPM) framework was used with some modifications (3). First, the denoising U-Net was modified so that it can take 100-channels input consisting of 50 dynamic scans of corrupted DSC MRI data as conditioning input and 50 generated denoised images. Second, the denoising U-Net was also enlarged where the convolution kernels were changed to 256, 512, 1024, 2048, and 4096 for each blocks respectively (figure 1). Other parameters used to train the DDPM were: diffusion steps = 1000, ADAM optimizer, batch size = 2, training epochs = 2500, and learning rate = 1e-4 with scheduler where learning rate was reduced when loss did not improve.
The generated DSC MRI data were evaluated against the corrupted DSC MRI data by visual comparison, ROI comparison, and brain mask DICE score.
Results
Representative generated DSC MRI compared against the corrupted DSC MRI data showed a successful generation of DSC MRI when the time points were corrupted by noise or movement (figure 2). The generated DSC MRI data also properly highlighted the vessel and gray matter structure throughout the contrast passage (figure 2, noised row). It should be noted that the contrast arrival and clearance timings were also generated properly even when some pre-contrast arrival time points were noised (figure 3a). The signal intensity in MCA and gray matter showed that the signal evolution of true DSC and generated DSC were closely matched. The generated DSC MRI also showed a more accurate brain shape as shown by the DICE score (figure 3b). The noised-corruption case showed worst DICE score due to the noised-time points attaining no resemblance to the DSC MRI. The movement corruption case showed the best DICE score compared to other corruption cases. Nevertheless, the DICE score from the generated DSC MRI were higher than any of the corrupted DSC MRI cases indicating accurate generation of brain structures in general.Discussion
The proposed generative diffusion model successfully restored the corrupted DSC MRI data while properly reflecting the time series and contrast passage time. There are two corruption cases described in this study which may not reflect all possible artifacts that may present in typical DSC MRI acquisition. However, the two corruption cases may be enough to test the ability of the generative diffusion model to restore both the pre-contrast images and contrast-enhanced images at proper timing. This method is not specific to DSC MRI data and may be extended to any dynamic MRI scans with further training.Conclusion
In this work, we proposed a generative diffusion model to restore corruption in DSC MRI data. The corrupted DSC MRI data were successfully corrected while the contrast arrival time and washout time were properly reflected. The brain shape comparison also showed close resemblance to the ground truth DSC MRI data. We believe the proposed approach can provide a solution to restoring a corrupted DSC MRI data while maintaining accurate contrast passage timing.Acknowledgements
No acknowledgement found.References
1. Duffy, Ben A., et al. "Retrospective motion artifact correction of structural MRI images using deep learning improves the quality of cortical surface reconstructions." Neuroimage 230 (2021): 117756.
2. Haskell, Melissa W., Stephen F. Cauley, and Lawrence L. Wald. "TArgeted Motion Estimation and Reduction (TAMER): data consistency based motion mitigation for MRI using a reduced model joint optimization." IEEE transactions on medical imaging 37.5 (2018): 1253-1265.
3. Ho, Jonathan, Ajay Jain, and Pieter Abbeel. "Denoising diffusion probabilistic models." Advances in Neural Information Processing Systems 33 (2020): 6840-6851.
Figures
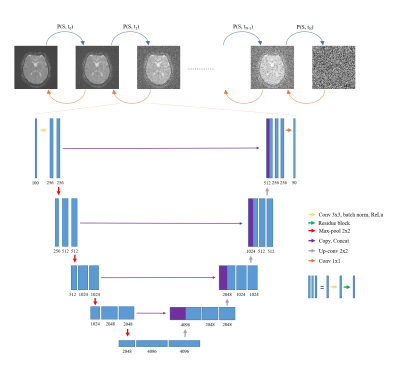
Figure 1. Denoising diffusion probabilistic model (DDPM) framework for
DSC MRI corruption correction. The denoising U-Net was modified so that it
takes 100 channel inputs and give 50 channel outputs. The convolution kernels
were also enlarged.

Figure 2. Comparison between three different corrupted DSC MRI cases and
its generated DSC MRI from generative diffusion model. The generative diffusion
model could correct for movement artifact, noised, and combined movement and
noised artifact. It can also properly generated post-contrast vessel and grey
matter structures.
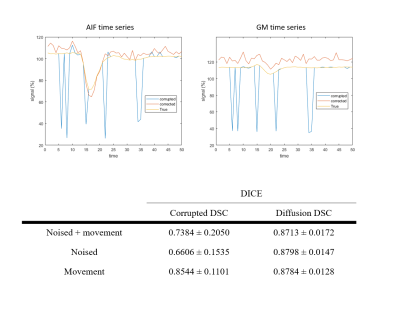
Figure 3. Representative time series of AIF and GM ROI (top row) from
noised + movement corrupted DSC MRI case showed a generative diffusion
model-generated DSC MRI time series (red line) follow closely to the true DSC
MRI time series (yellow line) compared to the corrupted DSC MRI time series
(blue line). The contrast arrival and washout timings of generated DSC MRI also
closely resemble that of true DSC MRI time series. The brain shape comparison
between corrupted DSC MRI and diffusion-generated DSC MRI also showed accurate
generation of brain structures in general.
DOI: https://doi.org/10.58530/2023/2946