2942
Automated hybrid approach to diagnose Parkinson's disease via deep learning and radiomics
Hongyi Chen1, Xueling Liu2, Yuxin Li1,2, Puyeh Wu3, and Daoying Geng1,2
1Academy for Engineering and Technology, Fudan University, Shanghai, China, 2Huashan Hospital, State Key Laboratory of Medical Neurobiology, Fudan University, Shanghai, China, 3GE Healthcare, Beijing, China
1Academy for Engineering and Technology, Fudan University, Shanghai, China, 2Huashan Hospital, State Key Laboratory of Medical Neurobiology, Fudan University, Shanghai, China, 3GE Healthcare, Beijing, China
Synopsis
Keywords: Data Analysis, Parkinson's Disease
In this study, we constructed a hybrid machine learning model utilizing CNN and radiomics features based on NM-sensitive setMag images. The hybrid features improved the diagnostic performance in distinguishing PD patients from HC, as demonstrated in the SVM classifier, which demonstrated 95.7% accuracy, 92.9% sensitivity, and 100% specificity. The interpretability of the radiomics approach is better because radiomics features provide more interpretable biomarkers, while the CNN approach extracts deeper features from images. Furthermore, visualizing regions that influence classification decisions via saliency map can also enhance the interpretability of the CNN approach.Background
Progressive loss of neuromelanin (NM) containing dopamine-producing neurons and increase in iron deposition in substantia nigra pars compacta (SNpc) are reliable and promising biomarkers for the diagnosis of Parkinson's disease (PD)1,2. Due to high iron and low NM content in SNpc, the diagnosis of PD patients can be made quickly by calculating the reflected signal intensity and contrast ratio (CR) on NM-sensitive MRI images3-5. However, these conventional methods rely on manual delineation of the ROI6,7, which is subjective, labor-intensive and error-prone. Recently, Takahashi et al.8 proposed an automatic voxel-based SNpc segmentation system, while it was subject to errors in the alignment process and insensitive to small SNpc regions.With the rapid development of deep learning and convolutional neural network (CNN), it has become a powerful tool for medical image applications9. However, there are several obstacles to the application of CNN in PD diagnosis. Due to the black-box nature of CNN models, this predisposes the diagnoses made by the models to be less interpretable. Secondly, CNNs that utilize image features cannot be effectively combined with histological features. Accordingly, researchers have started to focus on the combination of CNNs and radiomics. Choi et al.10 developed a fully automated model for segmentation and status prediction of gliomas by utilizing tumor MRI images and radiomics features, and achieved an satisfactory prediction accuracy in the external test.
In this study, NM-sensitive short-echo-time magnitude (setMag) MRI images11 were firstly reconstructed as inputs. We proposed a hybrid model with machine learning classifier, utilizing features extracted from CNN model and radiomics approach, and aimed to verify its feasibility and performance in PD diagnosis.
Methods
This study was approved by the Ethics Committee of our hospital (No. KY 2016–214). Informed consent was obtained from all participants. 73 PD patients and 48 healthy controls (HC) were recruited in this study. MRI examinations were performed on a 3.0-T MR scanner (MR750; GE Healthcare, Milwaukee, WI). 3D multi-echo GRE images were acquired, and NM-sensitive setMag images were reconstructed following the procedure published previously11. Bilateral hyperintensity ROIs in SNpc were manually depicted on setMag images using ITK-SNAP12.For the radiomics model, we extracted 107 radiomics features from original images using PyRadiomics, consisting of 6 categories: (1) first-order statistics; (2) GLCM; (3) GLDM; (4) GLRLM; (5) GLSZM; (6) NGTDM. Each original image was subjected to 7 transformations including: wavelet, LoG, square, square-root, logarithm, exponential, and gradient. In total, 1781 radiomics features were taken into consideration, and were further selected by LASSO to retain the most important features.
For the CNN model, YOLO v5 model was used to extract brainstem regions, and our CNN model was modified from the LeNet-513. Specifically, input images were sequentially passed through two sets of convolutional and maximum pooling layers, before being spanned into a vector in the final fully-connected layer.
Figure 1 shows entire process of the fusion and classification. In order to filter CNN features, we quantified the importance of features using the MDI evaluation method based on the random forest classifier. Top 20 features were selected and connected to the radiomics features. Subsequently, features were divided into training and validation sets at a ratio of 8:2. 7 algorithms were adopted including: k-nearest neighbor, support vector machine, random forests, logistic regression, Gaussian naïve Bayes, adaptive boosting and multilayer perceptron. Depending on the performance, multiple index system and classifier models were analyzed to determine the optimal model.
Results
The image set extracted by YOLO v5 model was fed into the CNN model for extracting features. After screening and fusion, we obtained a total of 121 cases with a feature vector of size 1 × 54 for each case. The results of the hybrid machine learning model on the validation set were shown in Table 1. In addition, we plotted ROC curves and calculated the AUC for each machine learning method (Figure 2). The best performing classifier of hybrid model was SVM: ACC = 0.957, SEN = 0.929, SPE = 1.000, PPV = 1.000, NPV = 0.900, F1-score = 0.963, and AUC = 0.99 (95% CI: 0.97–1.00), which was improved comparing to the radiomics model: ACC = 0.913, SEN = 0.929, SPE = 0.889, PPV = 0.929, NPV = 0.889, F1-score = 0.929, AUC = 0.94 (95% CI: 0.86–1.00).To explain the role of CNN features, we visualized the features extracted in the fully-connected layer of the CNN model. As shown in Figure 3, the focus of the saliency map was on the case characteristic "dovetail" of PD patients, which was consistent with the pathological change regions of PD patients in the current study and greatly increased the interpretability of our CNN model.
Conclusion
In this study, we proposed a hybrid machine learning model utilizing CNN and radiomics features for PD diagnosis. Our results demonstrated that this approach produced the desired outcome. Radiomics feature provided abundant texture information from images, and features extracted by CNN were continuously optimized based on the classification accuracy on the training set, enabling better problem solutions from data-driven perspective. Furthermore, according to the saliency map, different locations displayed varying degrees of significance which proves the interpretability of our hybrid classification model.Acknowledgements
No acknowledgement found.References
- Ward, R. J., Zucca, F. A., Duyn, J. H., Crichton, R. R. & Zecca, L. The role of iron in brain ageing and neurodegenerative disorders. The Lancet Neurology 13, 1045-1060 (2014).
- He, N. et al. Imaging iron and neuromelanin simultaneously using a single 3D gradient echo magnetization transfer sequence: Combining neuromelanin, iron and the nigrosome-1 sign as complementary imaging biomarkers in early stage Parkinson's disease. Neuroimage 230, 117810, doi:10.1016/j.neuroimage.2021.117810 (2021).
- Gaurav, R. et al. Longitudinal Changes in Neuromelanin MRI Signal in Parkinson's Disease: A Progression Marker. Mov Disord 36, 1592-1602, doi:10.1002/mds.28531 (2021).
- Fabbri, M. et al. Substantia Nigra Neuromelanin as an Imaging Biomarker of Disease Progression in Parkinson's Disease. J Parkinsons Dis 7, 491-501, doi:10.3233/JPD-171135 (2017).
- Prasad, S. et al. Three-dimensional neuromelanin-sensitive magnetic resonance imaging of the substantia nigra in Parkinson's disease. Eur J Neurol 25, 680-686, doi:10.1111/ene.13573 (2018).
- Taniguchi, D. et al. Neuromelanin imaging and midbrain volumetry in progressive supranuclear palsy and Parkinson's disease. 33, 1488-1492 (2018).
- Pyatigorskaya, N. et al. Magnetic Resonance Imaging Biomarkers to Assess Substantia Nigra Damage in Idiopathic Rapid Eye Movement Sleep Behavior Disorder. Sleep 40, doi:10.1093/sleep/zsx149 (2017).
- Takahashi, H. et al. Comprehensive MRI quantification of the substantia nigra pars compacta in Parkinson's disease. Eur J Radiol 109, 48-56, doi:10.1016/j.ejrad.2018.06.024 (2018).
- Le Berre, A. et al. Convolutional neural network-based segmentation can help in assessing the substantia nigra in neuromelanin MRI. Neuroradiology 61, 1387-1395, doi:10.1007/s00234-019-02279-w (2019).
- Choi, Y. S. et al. Fully automated hybrid approach to predict the IDH mutation status of gliomas via deep learning and radiomics. Neuro-oncology 23, 304-313 (2021).
- Liu, X. et al. Short-echo-time magnitude image derived from quantitative susceptibility mapping could resemble neuromelanin-sensitive MRI image in substantia nigra. 20, 262, doi:10.1186/s12883-020-01828-8 (2020).
- Yushkevich, P. A. et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31, 1116-1128 (2006).
- LeCun, Y. LeNet-5, convolutional neural networks. URL: http://yann. lecun. com/exdb/lenet 20, 14 (2015).
Figures
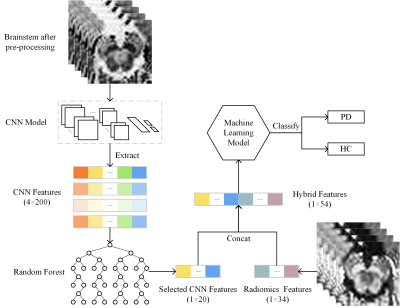
Figure 1. Flowchart
of classification based on CNN
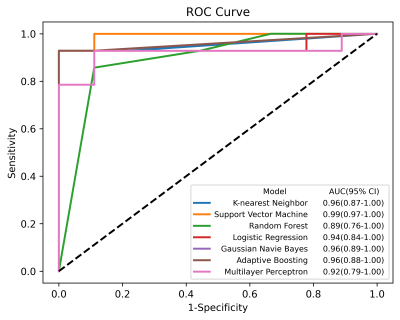
Figure 2. The
ROC curve of machine learning algorithms used hybrid features
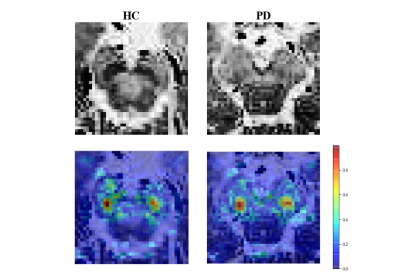
Figure 3. Saliency
map of the HC and PC case, with the original image in the upper half and the
resulting saliency map in the lower half.
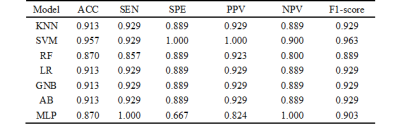
Table 1. The performance of each method, KNN ( k-nearest neighbor), SVM (support vector
machine), RF (random forests), LR (logistic regression), GNB (Gaussian naïve
Bayes), AdaBoost (adaptive boosting), MLP (multilayer perceptron), ACC
(accuracy), SEN (sensitivity), SPE (specificity), PPV (positive predictive
value), NPV (negative predictive value).
DOI: https://doi.org/10.58530/2023/2942