2941
Deep learning improves the estimation of fiber orientation distribution for tractography in the human1Radiology, NYU Langone health, new york, NY, United States, 2the State University of New York, Buffalo, NY, United States
Synopsis
Keywords: Visualization, Brain Connectivity, Diffusion MRI tractography
Although diffusion MRI (dMRI) tractography can map brain connectivity non-invasively, accurate tractography in the human brain remains challenging due to inherent and technical limitations. In this study, we demonstrate a deep learning (DL) based approach for improving the estimation of fiber orientation distribution (FOD) from dMRI data. Trained with augmented whole brain tractography results from high-resolution dMRI data, the DL approach outperformed conventional FOD estimation methods in crossing fiber regions with dMRI data at spatial and angular resolutions comparable to routine clinical scans. The approach can potentially shorten the dMRI acquisition necessary for accurate tractography and connectome analysis.Introduction:
Diffusion MRI (dMRI) tractography is the only non-invasive tool for mapping macroscopic structural connectivity in the brain but its ability to accurately reconstruct white matter pathways remains limited1,2. The estimation of fiber orientation distribution (FOD) from dMRI signals is critical because FODs direct the propagation of tractography streamlines, but complex intravoxel fiber configurations organization (e.g. crossing fibers) in certain brain regions can introduce uncertainties in FOD estimation3-5, which, despite progress in imaging acquisition and FOD estimation methods6-8, still pose a major obstacle for accurate reconstruction of axonal pathways in the brain.In this study, we investigated whether deep learning (DL) can improve FOD estimation in dMRI data, with the goal of achieving high-quality tractography within a clinically feasible time. We augmented whole brain streamlines obtained from the human connectome project (HCP) data with tract-specific tractography streamlines and used the augmented streamlines as targets to train DL networks to estimate FODs from dMRI data with spatial and angular resolutions comparable to routine clinical scans. Our results showed that the DL networks can improve FOD estimation in crossing fiber regions.
Methods:
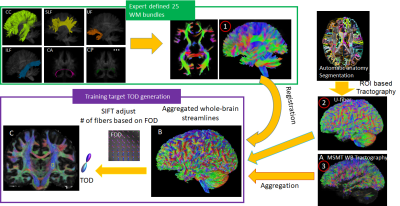
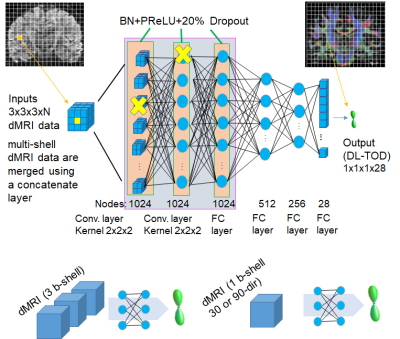
Whole brain and tract-specific tractography: FODs were estimated from 50 young adult human connectome project (HCP) dMRI data (1.25 mm resolution, 3 b-shells with 90 diffusion directions each) using multi-shell and multi-tissue (MSMT) constrained spherical deconvolution (CSD) method implemented in MRtrix6. Three sets of streamlined data were generated using the MSMT FODs: 1) 25 major white matter (WM) tracts fibers from the 2015 ISMRM Challenge (https://tractometer.org/ismrm2015/home/), reconstructed using the starting and ending voxels transformed to individual subject data in this study; 2) 30 cortical U-fibers, reconstructed based on individual cortical parcellation, as described in9; 3) Whole-brain tractography based on MSMT FOD (Fig. 1A). The three datasets were aggregated into an augmented whole brain streamline data (Fig.1B). Tract orientation distributions (TODs)10 of each subject were calculated after adjusting the number of streamlines using SIFT (Fig. 1C).Deep learning setup: A deep neural network based on 11 consisted of two convolutional and four fully connected layers, with a 20% drop-out in the first two layers. The input to the network was under-sampled HCP dMRI data (b=1,000 s/mm2 only, 30 or 90 direction, 1.25 or 2 mm resolution, with or without added Rician noises). The training targets were the augmented whole brain streamline TODs based on high-resolution dMRI data from the same subjects. Because the relationship between FODs and dMRI signals should be local and we would like to include minimal spatial information, we used small 3x3x3 voxel patches, instead of the entire image as input to the network, from 30 subjects’ brains for training (a total of three million voxels, 10% of which were used as validation) and 20 subjects for testing.
Results:
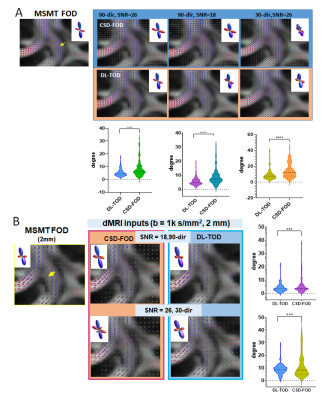
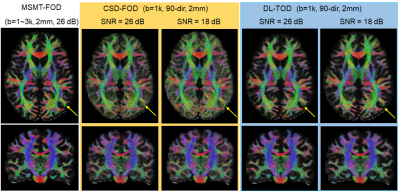
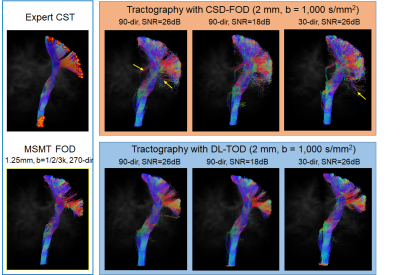
DL improved the estimation of FODs in crossing fiber regions: Using MSMT-FODs as ground truth, the peak orientations of network predicted TODs (DL-TODs) were compared with conventional CSD-FODs by measuring the average angle deviations between similar FOD peaks. For input dMRI data with a single b-shell and 90 directions, DL-TODs had significant reduced deviations from MSMT-FODs than CSD-FODs in the corona radiata(5.17°±2.7° v.s. 8.3°±6.1°, p<0.0001, n=20) (Fig. 3A). When noises were added to the dMRI data (SNR = 18 vs 26 in original), the orientation deviations became 5.62°±2.7° for DL-TOD v.s. 9.0°±5.5° for CSD-FOD in the corona radiata (p < 0.0001) (Fig. 3A). When only 30-direction data were used, the orientation deviation became 9.3°±6.6° for DL-TOD v.s. 14.2°±8.7° for CSD-FOD (p<0.0001). When the spatial resolution of the input data was reduced to 2 mm, DL-TOD still better preserved the orientation of MSMT-FOD than CSD-FOD (3.8°±2.6° (DL-TOD) v.s. 5.8°±6.1° (CSD-FOD) for 90-direction data with SNR =18 (p<0.0005); 8.8°±3.7° (DL-TOD) v.s. 12°±10.5° (CSD-FOD) for 30-direction with SNR = 26 (p<0.0005) (Fig. 3B).Improved tractography using DL-TODs: Whole brain tractography results using DL-TODs from low-resolution dMRI data (b=1,000 s/mm2, 90 direction) showed good agreement with results using MSMT-FOD, even with added noise. In comparison, tractography based on CSD-FODs showed more erroneous streamlines and small white matter tracts became less well-defined (Fig. 4). The corticospinal tract (CST) reconstructed from DL-TOD consistently showed reduced erroneous branches compared to results from CSD-FODs.
Discussions&Conclusion
Our results demonstrate that DL networks, trained with high-quality tractography results based on the HCP data, can enhance the accuracy of FOD estimation and tractography, particularly in regions with cross fibers, in dMRI data with lower spatial and angular resolutions. Similar to recent work by12, which used DL to accelerate diffusion tensor imaging, the technique developed here can potentially shorten the dMRI acquisition necessary for accurate tractography and connectome analysis in subjects that do not tolerate long acquisition.Due to scarce ground truth tract-tracing data from the human brain, the DL networks were trained using TODs from tractography streamlines from high-resolution HCP dMRI data. The addition of tract-specific tractography streamlines (large WM tracts and U-fibers) potentially adjusted the peak orientations along the paths of these tracts and increased the likelihood of these tracts being reconstructed, but the benefits were limited by the number of tracts included here.
Acknowledgements
No acknowledgement found.References
1. Thomas, C. et al. Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. P Natl Acad Sci USA 111, 16574-16579, doi:10.1073/pnas.1405672111 (2014).
2. Maier-Hein, K. H. et al. The challenge of mapping the human connectome based on diffusion tractography. Nat Commun 8, 1349, doi:10.1038/s41467-017-01285-x (2017).
3. Seehaus, A. K. et al. Histological validation of DW-MRI tractography in human postmortem tissue. Cereb Cortex 23, 442-450, doi:10.1093/cercor/bhs036 (2013).
4. Schilling, K. G. et al. Histological validation of diffusion MRI fiber orientation distributions and dispersion. NeuroImage 165, 200-221, doi:10.1016/j.neuroimage.2017.10.046 (2018).
5. Schilling, K. G. et al. Limits to anatomical accuracy of diffusion tractography using modern approaches. Neuroimage 185, 1-11, doi:10.1016/j.neuroimage.2018.10.029 (2019).
6. Jeurissen, B., Tournier, J. D., Dhollander, T., Connelly, A. & Sijbers, J. Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion MRI data. Neuroimage 103, 411-426, doi:10.1016/j.neuroimage.2014.07.061 (2014).
7.Tournier, J. D., Calamante, F. & Connelly, A. Robust determination of the fibre orientation distribution in diffusion MRI: non-negativity constrained super-resolved spherical deconvolution. Neuroimage 35, 1459-1472, doi:10.1016/j.neuroimage.2007.02.016 (2007).
8. Baete, S. H. et al. Fingerprinting Orientation Distribution Functions in diffusion MRI detects smaller crossing angles. Neuroimage 198, 231-241, doi:10.1016/j.neuroimage.2019.05.024 (2019).
9. Zhang, Y. J. et al. Atlas-guided tract reconstruction for automated and comprehensive examination of the white matter anatomy. Neuroimage 52, 1289-1301, doi:10.1016/j.neuroimage.2010.05.049 (2010).
10. Dhollander, T. et al. Track orientation density imaging (TODI) and track orientation distribution (TOD) based tractography. Neuroimage 94, 312-336, doi:10.1016/j.neuroimage.2013.12.047 (2014).
11. Lin, Z. et al. Fast learning of fiber orientation distribution function for MR tractography using convolutional neural network. Med Phys 46, 3101-3116, doi:10.1002/mp.13555 (2019).
12. Li, H. et al. SuperDTI: Ultrafast DTI and fiber tractography with deep learning. Magn Reson Med, doi:10.1002/mrm.28937 (2021).
Figures