2938
Accelerated MRI Reconstruction Using a Lightweight Recurrent Transformer: ReconFormer1Department of Computer Science, Johns Hopkins University, Baltimore, MD, United States, 2Department of Radiology, Johns Hopkins University, Baltimore, MD, United States
Synopsis
Keywords: Image Reconstruction, Image Reconstruction
Accelerating MRI reconstruction process is a challenging ill-posed inverse problem due to the excessive under-sampling operation in k-space. While state-of-the-art algorithms have shown a great progress based on convolutional neural networks (CNN), transformers for MRI reconstruction has not been fully explored in the literature. We propose a recurrent transformer model, namely, ReconFormer, for MRI reconstruction which can iteratively reconstruct high fertility magnetic resonance images from highly under-sampled k-space data. We validate the effectiveness of ReconFormer on multiple datasets with different magnetic resonance sequences and show that it achieves significant improvements over the state-of-the-art methods with better parameter efficiency.Target audience
Researchers and clinicians interested in MRI reconstruction methods.Purpose
Extending acquisition time to collect complete data in k-space imposes significant burden on patients and makes MRI less accessible. To accelerate MRI acquisition, one widely accepted approach, compressed sensing (CS), is capable of formulating the image reconstruction as solving an optimization problem with several assumptions including sparsity and incoherence [1]. Recently, advanced deep learning-based methods are gaining more attention for fast and accurate MRI reconstruction. However, CS algorithms and CNN-based MRI reconstruction have several deficiencies impeding their practicability in real-world applications. Transformer [2] models, introducing the self-attention mechanism to capture global interactions between contexts, open a new possibility to solve the challenging problem of MRI reconstruction. However, scale modeling is inefficient and inflexible. In this abstract, to overcome those issues, a novel Recurrent Transformer, termed ReconFormer, is proposed to recover the fully-sampled image from the under-sampled k-space data in accelerated MRI reconstruction.Methods
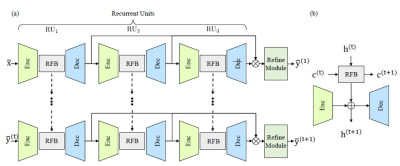
ReconFormer (Fig. 1) consists of three recurrent units and a refine module (RM). To maintain high-resolution information, ReconFormer employs globally columnar structure. In particular, recurrent units map the input degraded images to the original dimension. Meanwhile, across each recurrent unit, the receptive fields of recurrent units are gradually reduced to reconstruct high-quality MR images in a coarse-to-fine way. It is worth noting that the last recurrent unit RU3 employs overcomplete architecture [3], which has been demonstrated to efficiently constraint the receptive field. A recurrent unit (Fig. 1(b)) contains an encoder fEnc, a ReconFormer block fRFB, and a decoder fDec. To have a more stable optimization [4], encoder and decoder are built up on convolution layers. A data consistency layer is added at the end of each decoder network to reinforce the data consistency in the k-space. In addition, a ReconFormer block is formed by stacked recurrent pyramid transformer layers. The core design of RPTL is the Recurrent Scale-wise Attention (RSA). which operates on multi-scale patches in parallel. Such design enables efficient in-place scale modeling and forms a feature pyramid by projecting features at various scales directly into multiple attention heads. Consequently, the proposed RPTL allows scale processing at basic architecture units. In addition, the correlation estimation in the proposed RSA relies on both the hidden state and the deep feature correlation from the previous iteration, which enables more robust correlation estimation by propagating correlation information between adjacent states.The fastMRI [5] and HPKS [6] datasets are used for conducting experiments. The fastMRI dataset contains 1,172 complexvalued single-coil coronal proton density (PD)-weighted knee MRI scans. The HPKS dataset provides complex-valued single-coil axial T1-weighted brain MRI scans from 144 post-treatment patients with malignant glioma. In experiments, the input under-sampled image sequences are generated by randomly under-sampling the k-space data using the Cartesian under-sampling function that is the same as the fastMRI challenge [5]. Peak signal to noise ratio (PSNR) and structural index similarity (SSIM) are used as the evaluation metrics for comparison. We implement the proposed model using PyTorch on Nvidia RTX8000 GPUs.
To verify the effectiveness of the proposed ReconFormer, we compare the proposed method with 8 representative methods, including conventional compressed sensing (CS) based method [7], popular CNN-based methods – UNet [8], KIKI-Net [9], Kiu-net [10], and D5C5 [11], state-of-the-art iterative reconstruction approaches – PC-RNN [12] and OUCR [3], and vision transformer model –SwinIR [13].
Results and Discussion
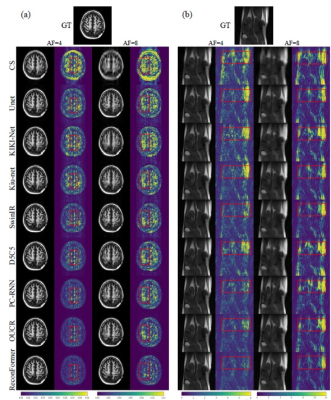
Table 1 shows the quantitative results evaluated on the two datasets for AF=4 and AF=8. Compared with the other methods, the proposed ReconFormer achieves the best performance on multiple datasets for all acceleration factors while containing the least number of parameters. It is worth noting that our method exhibits a larger performance improvement, when the acceleration factor increases (i.e., more challenging scenarios). In particular, for the HPKS and fastMRI datasets (Fig.2), our model outperforms the most competitive approach OUCR [8] by 0.9 dB and 0.3 dB in 8× acceleration, respectively. While the fastMRI dataset is more challenging due to the acquisition quality, all reported improvements achieved by ReconFormer are statistically significant.Conclusions
In this paper, we propose a recurrent transformer-based MRI reconstruction model ReconFormer. By leveraging the novel RPTL, we are able to explore the multi-scale representation at every basic building units and discover the dependencies of the deep feature correlation between adjacent recurrent states. ReconFormer is lightweight and does not require pre-training on large-scale datasets. Our experiments suggest the promising potential of using transformer-based models in the MRI reconstruction task.Acknowledgements
The authors thank our clinical collaborators for help with the patient recruitment and MRI technicians for assistance with MRI scanning. This study was supported in part by grants from the NIH.References
1. Haldar, J.P., Hernando, D., Liang, Z.P. IEEE transactions on Medical Imaging 30, 893–903 (2010).
2. Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A.N., Kaiser, Ł., Polosukhin, I. Advances in neural information processing systems. pp. 5998–6008 (2017).
3. Guo, P., Valanarasu, J.M.J., Wang, P., Zhou, J., Jiang, S., Patel, V.M. Medical Image Computing and Computer Assisted Intervention. pp. 13–23 (2021).
4. Xiao, T., Dollar, P., Singh, M., Mintun, E., Darrell, T., Girshick, R. Advances in Neural Information Processing Systems 34 (2021).
5. Knoll, F., et al. Radiology: Artificial Intelligence 2, e190007 (2020).
6. Jiang, S., et al. Clinical Cancer Research 25, 552–561 (2019).
7. Tamir, J.I., Ong, F., Cheng, J.Y., Uecker, M., Lustig, M. ISMRM Workshop on Data Sampling & Image Reconstruction (2016).
8. Ronneberger, O., Fischer, P., Brox, T. International Conference on Medical image computing and computer-assisted intervention. pp. 234–241 (2015).
9. Eo, T., Jun, Y., Kim, T., Jang, J., Lee, H.J., Hwang, D. Magnetic resonance in medicine 80, 2188–2201 (2018).
10. Valanarasu, J.M.J., Sindagi, V.A., Hacihaliloglu, I., Patel, V.M. International Conference on Medical Image Computing and Computer-Assisted Intervention. pp. 363–373 (2020).
11. Schlemper, J., et al. IEEE transactions on Medical Imaging 37, 491–503 (2017).
12. Wang, P., Chen, E.Z., Chen, T., Patel, V.M., Sun, S. arXiv:1912.00543 (2019).
13. Liang, J., Cao, J., Sun, G., Zhang, K., Van Gool, L., Timofte, R. IEEE International Conference on Computer Vision Workshops (2021).
Figures