2929
MixRecon: A neural network mixing CNN and Transformer utilizes hybrid representations of image features for Accelerated MRI Reconstruction1Department of Imaging and Interventional Radiology, The Chinese University of Hong Kong, Hong Kong, Hong Kong
Synopsis
Keywords: Image Reconstruction, Machine Learning/Artificial Intelligence
Accelerated reconstruction for magnetic resonance imaging (MRI) is a challenging ill-posed problem because of the excessive under-sampling operation in k-space. Existing CNN-based and Transformer-based solutions face difficulties obtaining powerful representation due to relatively unitary local or global feature modeling capability. In this study, we develop a dual-branch network that can simultaneously exploit the complementarity of the two-style features by leveraging the merits of CNN and Transformer, to generate high-quality reconstruction from zero-filled images in the spatial domain. Qualitative and quantitative results from the fastMRI dataset demonstrate that the proposed method can achieve improved performance compared with other benchmark methods.
Introduction
Magnetic resonance imaging (MRI) is an important technique in the field of medical imaging. Although MRI can provide superior soft tissue contrast as a noninvasive imaging modality, slow data acquisition is a major limitation1,2. To accelerate the scanning process, k-space under-sampling operations together with fast reconstruction methods have been explored. Recently, inspired by the success of deep learning, CNN and Transformer architectures have been developed for fast MRI reconstruction3-5 and demonstrate significant performance gain. Within CNN, the convolution operations are good at extracting local features but experience difficulty to capture global representations due to the limitation of its receptive field. Within vision transformer, the cascaded self-attention modules can fuse global representations among the compressed patch embeddings but unfortunately deteriorate local feature details. In this study, we designed a network termed MixRecon, mixing CNN and vision transformer structure to perform global and local modeling simultaneously for accelerated MRI reconstruction.Methods
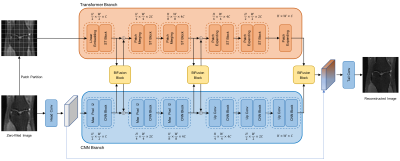
As shown in Figure 1, the proposed network, MixRecon, consists of two parallel branches: 1) CNN branch, which gradually increases the receptive field and encodes features from local to global using a UNet-like network. It takes under-sampled images as input. 2×2 max-pooling and up-convolution layer are respectively used for down-sampling in the left encoding part and up-sampling in the right decoding part. The CNN Block consists of two repeated 3×3 convolutions, each followed by a Rectified Linear Unit (RELU) layer and a BatchNorm (BN) layer. 2) Transformer branch, which starts with global self-attention and recovers the local details at the end using a Swin Transformer-like model. It takes non-overlapping image patches split by the patch partition module as input. A linear embedding layer is then applied to project the raw-valued feature to an arbitrary dimension (denoted as C). Similar to the CNN branch, patch merging and expanding modules are used for down-sampling and up-sampling in a patch-wise operation. The Swin Transformer (ST) block consists of a shifted window-based multi-head self-attention (MSA) module, followed by a 2-layer multi-layer perceptron (MLP) with Gaussian Error Linear Units (GELU) in between. A LayerNorm (LN) layer is applied before each MSA module and each MLP. Shallow, mid, and deep features with the same resolution extracted from both branches are fed into BiFusion Module to adaptively fuse the information. A Tail-Conv layer is utilized to generate the final reconstructed image.Experimental Setting
The proposed method is evaluated on the fastMRI6 single-coil knee dataset, which contains 1172 complex-valued single-coil coronal proton density (PD)-weighted knee MRI volumes with a matrix size of 320×320. We partition this dataset into 973 volumes for training, and 199 volumes (fastMRI validation dataset) for testing, with each volume roughly consisting of 36 slices. In the experiments, the input zero-filled images for training and testing are generated by applying Inverse Fourier Transform (IFT) to the under-sampled k-space data using the Cartesian under-sampling function released with the fastMRI dataset, with the acceleration factor of 4 and 8, respectively. Mean squared error (MSE) is used as the loss function in the training phase. Peak signal-to-noise ratio (PSNR) and structural similarity index measure (SSIM) are used as evaluation metrics for comparison.Results and Discussion
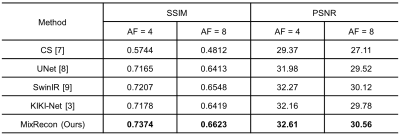
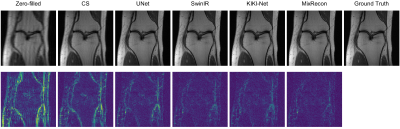
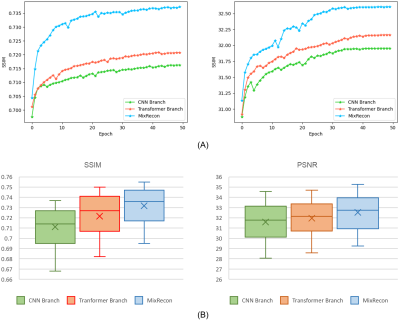
To evaluate the effectiveness of the proposed MixRecon, we compare MixRecon with four representative methods, including the conventional compressed sensing (CS) method7, popular CNN-based method - UNet8, vision transformer model - SwinIR9, and a commonly compared model - KIKI-Net3. The quantitative results of comparisons are shown in Table 1. Compared with the other methods, our proposed MixRecon achieves the highest SSIM and PSNR for both acceleration factors. Figure 2 shows the reconstructed MR images when acceleration factor (AF)=4. Note the obvious improvements of the results from our proposed MixRecon compared to the other methods. We also performed an ablation study to evaluate the effectiveness of network components – CNN branch and transformer branch. As shown in Figure 3, the proposed method has the best performance on the validation set compared to the CNN branch or the transformer branch alone, demonstrating the benefits of the hybrid features provided by the proposed dual-branch for reconstruction.Conclusion
In this paper, we propose a dual-branch network termed MixRecon to combine CNN with vision transformer for reconstruction of under-sampled MRI data. We demonstrated the proposed method can achieve superior results compared to the other benchmark methods using the fastMRI single-coil knee dataset. We expect this work can bring a new perspective on utilizing model-heterogenous features in MRI reconstruction tasks.Acknowledgements
This work is supported by a grant from the Innovation and Technology Commission of the Hong Kong SAR (Project MRP/001/18X), and a grant from the Faculty Innovation Award, the Chinese University of Hong Kong.References
1. Pal A, Rathi Y. A review and experimental evaluation of deep learning methods for MRI reconstruction. J Mach Learn Biomed Imaging. 2022; 1:001.
2. Zhu B, Liu JZ, Cauley SF, Rosen BR, Rosen MS. Image reconstruction by domain-transform manifold learning. Nature. 2018; 555(7697):487-492.
3. Eo T, Jun Y, Kim T, Jang J, Lee HJ, Hwang D. KIKI-net: cross-domain convolutional neural networks for reconstructing undersampled magnetic resonance images. Magn Reson Med. 2018; 80(5):2188-2201.
4. Huang J, Fang Y, Wu Y, et al. Swin transformer for fast MRI. Neurocomputing. 2022; 493: 281-304.
5. Zhou B, Schlemper J, Dey N, et al. Dsformer: A dual-domain self-supervised transformer for accelerated multi-contrast mri reconstruction. arXiv preprint arXiv:2201.10776, 2022.
6. Knoll F, Zbontar J, Sriram A, et al. fastMRI: A Publicly Available Raw k-Space and DICOM Dataset of Knee Images for Accelerated MR Image Reconstruction Using Machine Learning. Radiol Artif Intell. 2020; 2(1):e190007.
7. Lustig M, Donoho D L, Santos J M, et al. Compressed sensing MRI. IEEE signal processing magazine. 2008; 25(2): 72-82.
8. Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015; 234-241.
9. Liang J, Cao J, Sun G, et al. Swinir: Image restoration using swin transformer. Proceedings of the IEEE/CVF International Conference on Computer Vision. 2021; 1833-1844.
Figures