2905
Accelerated Brain ASL using 3D Gradient and Spin-Echo pseudo-Continuous ASL (GraSE-pCASL) with Compressed SENSE.
Yutaka Hamatani1, Kayoko Abe2, Masami Yoneyama3, Johannes M Peeters4, Kim van de Ven4, Michinobu Nagao2, Yasuhiro Goto1, Isao Shiina1, Kazuo Kodaira1, Takumi Ogawa1, Mana Kato1, and Shuji Sakai2
1Department of Radiological Services, Tokyo Women's Medical University Hospital, Tokyo, Japan, 2Department of Diagnostic imaging & Nuclear Medicine, Tokyo Women's Medical University Hospital, Tokyo, Japan, 3Philips Japan, Tokyo, Japan, 4Philips Healthcare, Best, Netherlands
1Department of Radiological Services, Tokyo Women's Medical University Hospital, Tokyo, Japan, 2Department of Diagnostic imaging & Nuclear Medicine, Tokyo Women's Medical University Hospital, Tokyo, Japan, 3Philips Japan, Tokyo, Japan, 4Philips Healthcare, Best, Netherlands
Synopsis
Keywords: Data Acquisition, Arterial spin labelling
In this study, 3D GraSE-pCASL was combined with Compressed SENSE (CS) to accelerate the acquisition time of perfusion images. The results showed that accelerated 3D CS-GraSE-pCASL could provide sufficient perfusion information in half the acquisition time compared to the conventional method. This technique may be useful in the diagnosis of cerebral blood flow disorders especially for pediatric patients and/or patients with cognitive impairments.Introduction
3D ASL does not require the administration of exogenous contrast agents, which is contraindicated in renal failure, to perform noninvasive quantification of cerebral blood flow (CBF) in absolute physiological units.1 The utility of 3D ASL for characterizing perfusion in both acute stroke and chronic cerebrovascular disease has been demonstrated, and the ability to quantify CBF should be useful for monitoring the effects of disease progression and therapy in patients with hypoperfusion syndromes, including pediatric populations.2.3 3D ASL is also sensitive to local CBF and metabolic changes that occur in degenerative diseases such as epilepsy and Alzheimer's disease.4 One of the challenges of 3D ASL in clinical practice is its long acquisition time which potentially has a negative impact especially for pediatric patients or patients with cognitive impairments, resulting in reduced accuracy due to the misregistration between control and label images caused by motion during the scan. Recently, a combination of sensitivity encoding (SENSE) and compressed sensing technique (Compressed SENSE) has been developed to reduce the acquisition time while maintaining the image quality. The reduction in acquisition time with CS has been demonstrated with various MRI techniques, including 2D EPI-based ASL.5.6,7 In this study, we attempted to develop the accelerated 3D ASL sequence combined with Compressed SENSE reconstruction.Methods
Five healthy volunteers (4 male, 1 female, age 25-45 years old) were examined by 3.0T MRI (Ingenia, Philips Healthcare) and dS-head coil. The study was approved by the local IRB (Clinical Trial Review Board) and all subjects gave written informed consent. ASL was performed by 3D Grase-pCASL. The parameters were: FOV=240x240x84 mm, slice number=14, Voxel size=3.75×3.75×6.0mm, Matrix=64×60×14, slice thickness=6.0 mm, TR/TE =4250/12ms, Flip angle 90°, Fat suppression=SPIR, pCASL label duration =1800ms, post label delay (PLD)=2000ms, acquisition time=5min. SENSE and CS factors were set to 1.4, 2.2, and 4.3 with corresponding acquisition times of 3:50, 2:20, and 1:40 minutes, respectively. We evaluated the consistency between 3D GraSE-pCASL without acceleration and with SENSE and CS acceleration. Six regions of interest (ROI) in the perfusion images were set up in the left and right gray matter (GM) at the level of the basal ganglia. Mean signal intensity (SI) and standard deviation (SD) were obtained over these ROIs. We used SI and SD to perform relative standard deviation (RSD) comparisons and Bland-Altman analysis. RSD was calculated as follows: RSD=SD/SI(Average). In the Bland-Altman analysis, P values <0.05 were considered significant different.Results and Discussion
Perfusion images and color maps of the 3D GraSE-pCASL were shown in the Figures 1 and 2. Figure 3 showed the results of the RSD analysis. SENSE showed variation in RSD, but CS had no variation in RSD. Figures 4 and 5 provide the results of Bland-Altman analysis of 3D GraSE-pCASL perfusion images. There was significant discordance between 3D GraSE-pCASL with SENSE and 3D GraSE-pCASL without SENSE. There was no significant discordance between 3D GraSE-pCASL with CS and 3D GraSE-pCASL without CS. Variation in SI was observed for CS factor 4.3, but not for CS factor 1.4 and 2.2. The reason for the RSD variation and SI discrepancy in SENSE was likely due to the presence of intense noise throughout the image. On the other hand, the lack of SD variation and SI discrepancy may be a result of the inherent denoising with CS. The fact that the agreement was maintained, even at high CS factors, suggests that 3D GraSE-pCASL has high signal sparsity allowing to undersample and accelerate by CS. It is shown that 3D GraSE-pCASL with CS is able to provide accurate perfusion information with high reproducibility and robustness in a short acquisition time.Conclusion
Accelerated 3D CS-GraSE-pCASL could provide sufficient perfusion information in half the acquisition time compared to the conventional method. This technique may be useful in the diagnosis of cerebral blood flow disorders especially for pediatric patients and/or patients with cognitive impairments.Acknowledgements
No acknowledgement found.References
1. Luis Hernandez-Garcia, rt al. Recent progress in ASL. Neuroimage. 2019 Feb 15;187:3-16. 2. André Monteiro Paschoal, et al. Contrast-agent-free state-of-the-art MRI on cerebral small vessel disease-part 1. ASL, IVIM, and CVR. NMR Biomed. 2022 Aug;35(8):e4742. 3. Xavier Golay et al. Multidelay ASL of the pediatric brain. Br J Radiol. 2022 Jun1;95(1134):20220034. 4. Avinash Chandra, et al. Magnetic resonance imaging in Alzheimer's disease and mild cognitive impairment. J Neurol. 2019 Jun;266(6):1293-1302. 5. Michael Lustig, et al. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn Reson Med. 2007 Dec;58(6):1182-95. 6. Sairam Geethanath, et al. Compressed sensing MRI: a review. Crit Rev Biomed Eng. 2013;41(3):183-204. 7. Kosuke Morita, et al. Rapid Brain Arterial Spin Labeling (ASL) using pseudo-Continuous ASL Echo Planar Imaging with Compressed SENSE (pCASL-EPICS). Proc. ISMRM. 2020:1621Figures
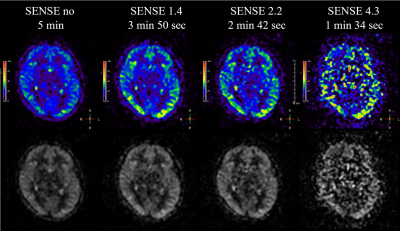
Figure.1
Perfusion images and color maps for
different SENSE factors are shown. As the SENSE factor increased, noise caused
signal irregularities.
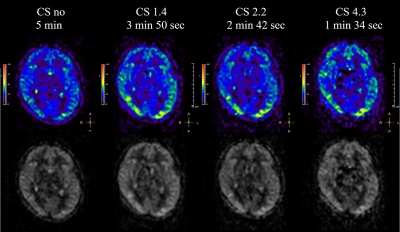
Figure.2
Perfusion images and color maps for
different CS factors are shown. The reproducibility of the perfusion images is
maintained even when the CS factor is increased.
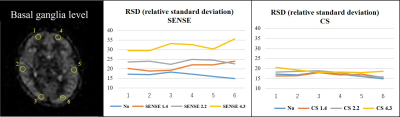
Figure.3
RSD results for one volunteer are shown.
SENSE showed greater variability in RSD with higher SENSE factor. CS had low
RSD variability even at high CS factors.
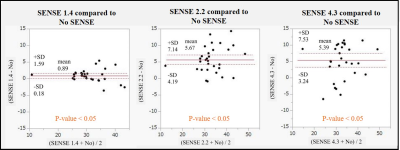
Figure.4 Consistency Assessment between 3D
GraSE-pCASL without SENSE and 3D GraSE-pCASL with SENSE using Brand-Altman
analysis by 5 volunteers. SENSE was less consistent compared to the reference
images without SENSE.
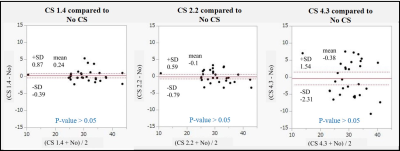
Figure.5
Consistency Assessment between 3D
GraSE-pCASL without CS and 3D GraSE-pCASL with CS using Brand-Altman analysis
by 5 volunteers. CS was highly congruent with the results obtained without
acceleration.
DOI: https://doi.org/10.58530/2023/2905