2898
Prediction of Genomic Signature of Prostate Lesion Radiosensitivity by mpMRI Radiomics and Machine Learning1University of Miami, Miami, FL, United States
Synopsis
Keywords: Radiomics, Prostate, multi-parametric MRI, prostate cancer radiosensitivity, genomic siganture, PORTOS
Genomic classifiers, such as PORTOS, have shown great promise in the prediction of prostate cancer radiosensitivity. However, the spatial heterogeneity of prostate cancer may confound genomic assessment due to tumor sampling error. We aimed to develop a model predictive of PORTOS genomic signature using multiparametric MRI (mpMRI) radiomics features and machine learning. Lesions were localized based on Habitat Risk Score maps. Eight radiomic features were selected (out of 167) including T2, ADC, high B-value intensity and texture variables and used to build logistic regression models through cross-validation. Our analysis shows association between the radiomics profile and prostate lesion radiosensitivity phenotype.Background and aim
Radiotherapy (RT) plays an important role in the treatment of prostate cancer, but its therapeutic effectiveness is highly variable. Genomic classifiers are a promising tool towards improved lesion characterization and patient risk stratification. PORTOS (Post-Operative Radiation Therapy Outcomes Score) is a 24-gene signature developed to predict which patients would benefit most from RT 1. Patients with a high PORTOS (>0.8) are less likely to develop metastasis at 10 years following RT than those who did not receive RT 1. However, similarly to pathology evaluation, the spatial heterogeneity of prostate cancer may confound genomic assessment due to tumor sampling error. The aim of this study is to develop a model predictive of PORTOS genomic signature of prostate lesion radiosensitivity using multiparametric MRI (mpMRI) radiomics features and machine learning.Methods
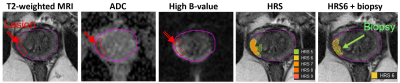
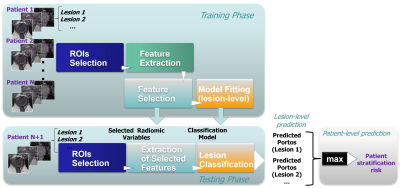
Patients have undergone mpMRI, followed by MRI-ultrasound (MRI-US) fusion biopsy as a part of clinical trials at University of Miami. Lesions were localized based on the Habitat Risk Score (HRS) maps derived from perfusion and diffusion MRI 2 and radiomic features were extracted from HRS=6 volume (Figure 1). PORTOS was assessed using gene expression analysis of the biopsy tissue, sampled also based on HRS. A multireference normalization approach was utilized for T2-weighted MRI intensities normalization 3. Specifically, a deep neural network (Mask-RCNN) was trained for automatic segmentation of three reference contours, namely the gluteus maximus (GM), femoral head and bladder 4. Then for each patient, a linear fit was estimated that mapped the average intensity values within these reference contours to corresponding reference values (defined before-hand). Six intensity and texture features were extracted from T2, ADC and high B-value images 5 and summarized through 9 statistical parameters. In addition, 4 parameters from Dynamic-Contrast Enhanced (DCE) MRI (time of contrast onset, Ktrans, kep, and ve) and the HRS6 volume were incorporated, resulting in 167 variables in total. All variables were scaled in [0, 1] range before subsequent analysis. Feature ranking was performed using the Maximum Relevance – Minimum Redundancy (MRMR) algorithm 6, followed by exhaustive feature selection among the top-15 ranked features. The feature subset selected as best was obtained by maximizing the area under the receiver operating characteristic curve (ROC-AUC) within a stratified 5-fold cross validation setting. Subsequently, 5-fold cross-validation (across patients) was used to train and evaluate logistic regression models for prediction of lesion radiosensitivity. To account for class imbalance, data were augmented by synthesizing new examples from the minority class (within the training set at each of the 5 folds) using the Synthetic Minority Over-sampling Technique (SMOTE) 7. Patient risk stratification was performed by selecting for each patient the lesion with the highest predicted PORTOS. The overall prediction modeling framework is illustrated in Figure 2.Results
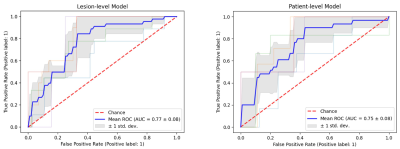
A total of 231 lesions across 78 patients were analyzed. Thirty-two of the analyzed lesions had high PORTOS (>0.8) and 21 patients with multiple lesions had both low and high PORTOS lesions. Eight radiomic features were selected and used to build the classification model. The average (across the testing sets) ROC-AUC was 0.77 ± 0.08 for lesion-level prediction and 0.75 ± 0.08 for patient-level prediction (Figure 3). The classification accuracy, at the default threshold of 0.5 for the prediction scores, was 0.73 for lesion-level prediction, and the average between sensitivity and specificity was 0.70.Conclusions
To the best of our knowledge, this is the first study to model the PORTOS gene signature with in vivo mpMRI radiomics. The analysis indicates that there is association between the radiomics profile and tumor radiosensitivity phenotype. Developing robust models with machine learning will allow for characterizing areas in the prostate with high radiation sensitivity and these spatial maps may serve as targets for the delivery of adjusted dose based on generated isotoxic plans optimized for dose escalation/tumor control. The incorporation of the clinical features and DCE temporal patterns in the machine learning framework will potentially improve the predictive power of the model.Acknowledgements
No acknowledgement found.References
1. Zhao SG, Chang SL, Spratt DE, et al. Development and validation of a 24-gene predictor of response to postoperative radiotherapy in prostate cancer: a matched, retrospective analysis. Lancet Oncol. 2016;17(11):1612-1620. doi:10.1016/S1470-2045(16)30491-0
2. Stoyanova R, Chinea F, Kwon D, et al. An Automated Multiparametric MRI Quantitative Imaging Prostate Habitat Risk Scoring System for Defining External Beam Radiation Therapy Boost Volumes. Int J Radiat Oncol Biol Phys. 2018;102(4):821-829. doi:10.1016/j.ijrobp.2018.06.003
3. Stoilescu L, Maas MC, Huisman HJ. Feasibility of multireference tissue normalization of T2-weighted prostate MRI. In Proceedings of the 34th annual scientific meeting, European Society for Magnetic Resonance in Medicine & Biology, Barcelona 2017 (Vol. 353).
4. Breto AL, Zavala-Romero O, Xu IR, et al. Deep Learning Approach for Multi-Reference Tissue Normalization on T2-weighted MRI in Longitudinal Dataset from Prospective Radiotherapy Trial for Prostate Cancer. International Journal of Radiation Oncology*Biology*Physics. 2020;108(3):S130. doi:10.1016/j.ijrobp.2020.07.858
5. Kwon D, Reis IM, Breto AL, et al. Classification of suspicious lesions on prostate multiparametric MRI using machine learning. J Med Imaging (Bellingham). 2018;5(3):034502. doi:10.1117/1.JMI.5.3.034502
6. Zhao Z, Anand R, Wang M. Maximum relevance and minimum redundancy feature selection methods for a marketing machine learning platform. In2019 IEEE international conference on data science and advanced analytics (DSAA) 2019 Oct 5 (pp. 442-452). IEEE. doi: 10.1109/DSAA.2019.00059.
7. Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority over-sampling technique. Journal of artificial intelligence research. 2002 Jun 1;16:321-57. doi: 10.1613/jair.953
Figures