2888
Comparison of radiomics-based machine learning survival models in predicting prognosis of glioblastoma1China-Japan Friendship Hospital, Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China, 2Department of Radiology, Liaocheng People's Hospital, Shandong First Medical University & Shandong Academy of Medical Sciences, Liaocheng, China, 3China-Japan Friendship Hospital, Beijing, China
Synopsis
Keywords: Radiomics, Cancer
In this study, we aimed to compare the performance of radiomics-based machine learning survival models in predicting the prognosis of glioblastoma multiforme (GBM) patients. The Cox proportional-hazards model (Cox-PH) and SurvivalTree, Random survival forest (RSF), DeepHit, DeepSurv four machine learning models were constructed, and the performance of the models was evaluated using C-index. We found that deep learning algorithms based on radiomics in predicting the overall survival of GBM patients, and the DeepSurv model showed the best predictive ability.
Introduction
Glioblastoma multiforme (GBM), as the most common malignant tumor in the central nervous system, nowadays has the poorest prognosis [1]. Machine learning is a branch of artificial intelligence that has a wide range of applications in diagnosing and prognostic assessing GBM [2]. The Cox proportional-hazards model (Cox-PH) is the most well-known method used to determine the association between clinical predictor variables and the risk of mortality events. Compared to CPH models, machine learning can identify clinically significant risks with some marginal variables that can significantly improve the model's performance. The Deepsurv model is a deep learning technique applied to a non-linear cox proportional risk network [3]. To our knowledge, no study has been conducted on the prognosis of GBM patients using radiomics combined with machine learning. Therefore, this study aimed to construct a traditional CPH model, a tree-based SurvivalTree model, an RSF model based on ensemble learning, a DeepSurv and DeepHit model based on deep learning for predicting the overall survival of GBM patients based on GBM radiomics data and clinical data and to compare the performance of the five models.Materials and Method
Patients data collectionA total of 131 patients with GBM were subsequently retrospectively enrolled in our study. Fluid attenuated inversion recovery (FLAIR) images and clinical information of corresponding patients were collected from The Cancer Imaging Archive (TCIA) datasets. The flow chart for this study is shown in Figure 1.
Construction of the radiomics signature
Using ITK-SNAP (www.itk-snap.org) software to segment the FLAIR images of patients in 3D. The least absolute shrinkage and selection operator (LASSO) method was used to select key features from the dataset significantly associated with prognosis. The selected features were linearly combined according to their respective coefficient weights to construct a radiomics signature.
Construction of the model
For the CPH model, proportional risk assumptions were made using the CoxPHFitter function. SurvivalTree model is based on the tree structure, and the tree building mainly includes tree generation and pruning. Random Survival Forest (RSF) is a combination of random forest and survival analysis methods. DeepSurv is a feed-forward deep neural network for CPH models to model a nonlinear representation of the risk of clinical events based on input features. The DeepHit model was initially designed to analyze the competing risks of multiple events.
Data analysis
After data preprocessing, the data from the training set was modeled by dividing the data into 70% training data and 30% test data. For the SurvivalTree and RSF models, the hyperparameters of the models were searched for using cross-validation. For the DeepSurv and DeepHit models, the hyperparameters of the models were searched for using manual optimization. Finally, the performance of the models was compared using Harrell's concordance index (C-index) and brier scores. The overall estimate of the brier score for all available times is called the Integrated Brier Score (IBS).
Results
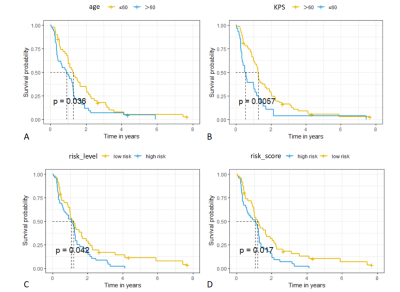
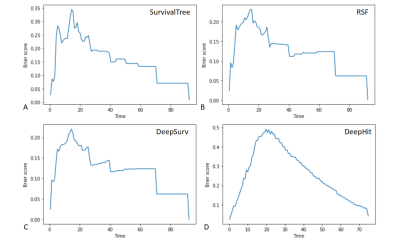
For the CPH model, the univariate cox analysis showed that age, Karnofsky performance status (KPS) score, radiomics risk score, and radiomics risk level were prognostic factors for overall survival in GBM. The KM survival curves for variables that were significant for the univariate survival analysis are shown in Figure 2. The C-index of the model was divided into 0.663 and 0.635 in the training and test set, and the IBS was 0.102 (Figure 3).For the SurvivalTree, the C-index of the model was divided into 0.702 and 0.655 in the training and test set, and the IBS was 0.192. For the RSF model, the C-index of the model was divided into 0.735 and 0.667 in the training and test set, and the IBS was 0.152 (Figure 3).
For the DeepSurv model, the C-index of the model was divided into 0.882 and 0.732 in the training and test set, and the IBS was 0.116. For the DeepHit model, the C-index of the model was divided into 0.608 and 0.560 in the training and test set, and the IBS was 0.261 (Figure 3).
Discussion and Conclusion
Precision treatment of GBM can slow down tumour growth and help improve patient prognosis. Previous studies on GBM have used deep learning for diagnostic and prognostic assessment of tumours [4]. To our knowledge, this is the first study to use machine learning and radiomics approaches to assess the prognosis of GBM patients. In this study, by constructing radiomics prognostic labels, using different machine learning models and comparing the performance with the traditional CPH model, the results show that the DeepSurv deep learning model shows superior predictive power compared to the traditional CPH model.In conclusion, based on the TCIA databases combined with a radiomics approach, this study confirmed that the DeepSurv model based on deep learning exhibited higher performance in GBM patient data compared with the CPH model. Based on the above-optimized model, a personalized treatment recommendation system for GBM can be developed to predict patient prognosis accurately.
Acknowledgements
The authors thank Dr. Lizhi Xie from GE Healthcare for help in solving MR technical problems.References
1. Ostrom Q T, Patil N, Cioffi G, et al. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System tumours Diagnosed in the United States in 2013-2017[J]. Neuro Oncol, 2020, 22(12 Suppl 2): iv1-iv96.
2. Lao J, Chen Y, Li Z C, et al. A Deep Learning-Based Radiomics Model for Prediction of Survival in Glioblastoma Multiforme [J]. Sci Rep, 2017, 7(1): 10353.
3. Katzman J L, Shaham U, Cloninger A, et al. DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network [J]. BMC Med Res Methodol, 2018, 18(1): 24-27.
4. Moradmand H, Aghamiri S M R, Ghaderi R, et al. The role of deep learning-based survival model in improving survival prediction of patients with glioblastoma[J]. Cancer Med, 2021, 10(20): 7048-7059.
Figures