2884
Prediction of overall survival for astrocytoma with whole-tumor radiomics analysis based on diffusion kurtosis imaging
Yan Tan1, Dawei Tian1, Wenqiao Zheng1, Xiaochun Wang1, and Hui Zhang1
1Department of Radiology, First Hospital of Shanxi Medical University, Taiyuan, China
1Department of Radiology, First Hospital of Shanxi Medical University, Taiyuan, China
Synopsis
Keywords: Radiomics, Diffusion/other diffusion imaging techniques
For investigating the usefulness of radiomics signature based on diffusion kurtosis imaging in overall survival prediction for astrocytoma, radiomics features extracted from whole-tumor on MK and MD images were selected to construct radiomics signature. Then the radiomics signature was integrated with clinical-genetic risk factors to develop a combined model, which was represented as nomogram. The results showed that the radiomics signature have offered clinically relevant prognostic information for astrocytoma and further stratified patients into different risk groups. The combined model achieved the highest predictive performance and facilitated clinical decision-making by nomogram, demonstrating the incremental value of radiomics signature.Background
Astrocytomas are the most common primary brain malignancy with variable morphologies, molecular subtypes, and clinical outcomes [1]. Diffusion kurtosis imaging (DKI) is a novel technique for quantizing the non-Gaussian water diffusion in brain tissues, which characterizes the degree of diffusion limitation and the inhomogeneity of diffusion, reflecting the complexity and heterogeneity of microstructure [2; 3]. The parameters provided by DKI mainly include mean kurtosis (MK) and mean diffusivity (MD) [4]. Currently, DKI has demonstrated that MK and MD are significant predictors of overall survival (OS) based on conventional parameters values [5-8]. The purpose of our study is to evaluate the prognosis of astrocytoma using radiomics signature of MK and MD sequences based on machine learning, clinical risk factors and molecular markers were further combined to predict OS of patients and guide clinical treatment accurately.Methods
We collected 58 patients with the diagnosis of WHO grade 2-4 astrocytoma. The clinical data, preoperative MRI images, follow-up time, and tumor genotypes including isocitrate dehydrogenase (IDH), O6-methylguanine-DNA methyltransferase (MGMT), telomerase reverse transcriptase (TERT), and deletion of 1p/19q were obtained.The tumors were initially segmented by one radiologist with 10-year experience using ITK-SNAP (http://www.itksnap.org) and determined by another radiologist with 15-year experience. We extracted radiomics features including shape features, first-order features, texture features and wavelet filtered features from the whole-tumor region of interest (ROI) for MK and MD images. Using the least absolute shrinkage and selection operator (LASSO) Cox regression and Pearson correlation analyses, three radiomics signatures (referred as Radioscore) were constructed by the selected most valuable prognostic features.
The radiomics signature divided astrocytoma patients into high-risk and low-risk groups based on the cut-off value calculated by the X-tile software [9]. Comparison of survival in the two groups was analyzed by Kaplan-Meier survival curve and log-rank test. In addition, the effectiveness of the radiomics signatures in predicting 1-, 2-, and 3-year survival was evaluated using the Receiver-Operating-Characteristic (ROC) curve and area under the curve (AUC). Univariate and multivariate Cox regression analyses were performed to obtain independent clinical and genetic variables and establish the clinical-genetic model. After that, the patients were stratified into various subgroups according to statistically significant clinical-genetic risk factors to further evaluate the incremental value of optimal radiomics signature for stratified analysis.
Then, the clinical and genetic risk factors corresponding to clinical-genetic model and the radiomics signature were used to construct the combined model, graphically presented as a nomogram. The consistency between the predicted and the actual probability was evaluated using the calibration curve. Decision curve analysis (DCA) was performed to estimate the clinical usefulness of the clinical-genetic model, radiomics signature, and the combined model. Finally, the generalization of the combined model was explained using k-fold (k=5) internal cross-validation.
Results
From MK and MD images, 851 radiomics features were extracted respectively for this study. The inter-observer interclass correlation coefficient of radiomics features ranged from 0.856 to 0.976, indicating good inter-observer consistency on ROI segmentation. There were 5, 5, and 6 radiomic features form the whole-tumor for MK, MD and combined sequences, respectively.According to the cut-off value of radiomics signatures, patients were classified into high-risk and low-risk groups. The radiomics signature of combined sequences performed greater AUC (0.72/0.82/0.84) than that of MD (0.7/0.8/0.82), and MK (0.66/0.7/0.79) respectively in predicting 1-, 2-, and 3-year OS (Figure.1). The radiomics signature of combined sequences (0.733, 95% CI: 0.688-0.779, P<0.001) performed better than that of MD (0.717, 95% CI: 0.670-0.764, P<0.001) and MK (0.696, 95% CI: 0.643-0.749, P<0.001) in terms of C-index.
Multivariate Cox regression analyses showed that grade, treatment, and MGMT were independent risk factors for OS, with a C-index of 0.761 (95% CI: 0.714-0.808) for the constructed clinical-genetic model. The patients were stratified by grade, treatment, and MGMT to form high-grade (3-4) astrocytoma group and low-grade (2) astrocytoma group, surgery group and combination therapy group, non-MGMTmet group and MGMTmet group respectively. The patients in these subgroups were further stratified into high-risk (Radioscore≥0.053) and low-risk (Radioscore<0.053) groups by radiomics signature except for non-MGMTmet group (Figure.2). The images of high-risk and low-risk patients with high-grade as well as low-grade astrocytoma are illustrated in Figure.3 and 4.
The combined model integrating the radiomics signature, grade, treatment, and MGMT performed a C-index of 0.832 (95% CI: 0.797-0.866) and was visualized by nomogram (Figure.5A). The calibration curves of OS for 1, 2, and 3 years exhibited favorable consistency between the nomogram predicted and actual survival probability (Figure.5B). The DCA revealed that the combined model achieved a greater clinical net benefit than radiomics signature and clinical-genetic model throughout the majority of the range of threshold probabilities (Figure.5C). The patients were randomly split into five parts for the internal cross-validation, which validated the combined model with an average C-index of 0.804.
Conclusions
In conclusion, our radiomics signature based on DKI was a prognostic imaging biomarker of astrocytoma for evaluating OS, and could stratify the prognosis of patients over the clinical-genetic risk factors. The combined model that integrating radiomics signature, grade, treatment, and MGMT improved the performance of clinical-genetic model and facilitated clinical decision-making by nomogram, demonstrating the incremental prognostic value for radiomics signature.Acknowledgements
This study has received funding by the National Natural Science Foundation of China (U21A20386, 81971593 to Hui Zhang; 82071893 to Yan Tan; 81971592 to Xiao-Chun Wang); the Youth Innovation Fund (YC1426 to Yan Tan).References
[1] Jiang T, Mao Y, Ma W et al (2016) CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett 375(2):263-273[2] Jiang R, Jiang J, Zhao L et al (2015) Diffusion kurtosis imaging can efficiently assess the glioma grade and cellular proliferation. Oncotarget 6(39):42380-42393
[3] Steven AJ, Zhuo J, Melhem ER (2014) Diffusion kurtosis imaging: an emerging technique for evaluating the microstructural environment of the brain. AJR Am J Roentgenol 202(1): W26-33
[4] Van Cauter S, Veraart J, Sijbers J et al (2012) Gliomas: diffusion kurtosis MR imaging in grading. Radiology 263(2):492-501
[5] Zhang J, Jiang J, Zhao L et al (2019) Survival prediction of high-grade glioma patients with diffusion kurtosis imaging. Am J Transl Res 11(6):3680-3688
[6] Wang X, Li F, Wang D, Zeng Q (2020) Diffusion kurtosis imaging combined with molecular markers as a comprehensive approach to predict overall survival in patients with gliomas. Eur J Radiol 128:108985
[7] Wang X, Gao W, Li F, Shi W, Li H, Zeng Q (2019) Diffusion kurtosis imaging as an imaging biomarker for predicting prognosis of the patients with high-grade gliomas. Magn Reson Imaging 63:131-136
[8] Li Y, Kim MM, Wahl DR, Lawrence TS, Parmar H, Cao Y (2021) Survival Prediction Analysis in Glioblastoma With Diffusion Kurtosis Imaging. Front Oncol 11:690036
[9] Camp RL, Dolled-Filhart M, Rimm DL (2004) X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 10(21):7252-7259
Figures
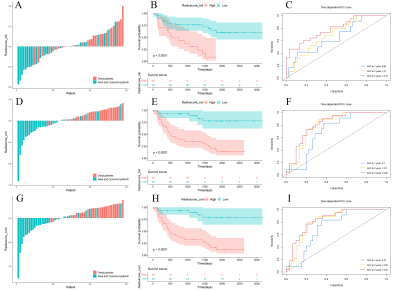
Figure 1 The validation of radiomics signatures for MK (A-C), MD (D-F), and combined sequences (G-I). In the specific distribution of Radioscore, red histograms represent death, and blue histograms represent patients with live or censored status (A, D, and G). The Kaplan-Meier survival curves of radiomics signatures for MK (B), MD (E), and combined sequences (H). ROC curves predicting 1-year, 2-year, and 3-year survival validated the great prognostic value of the radiomics signatures (C, F, I).
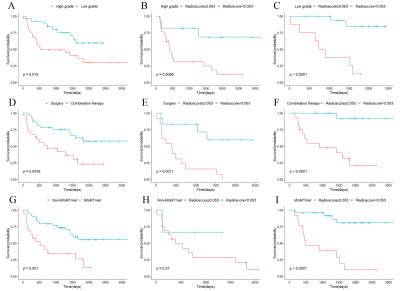
Figure 2 Stratified analyses of grade, treatment, and MGMT based on radiomics signature. The Kaplan-Meier plots illustrate overall survival of astrocytoma patients among different WHO grades (A), treatment (D), and MGMT methylation status (G). The patients were further stratified according to the cut-off value of radiomics signature among subgroups for high grade (B), low grade (C), surgery (E), combination therapy (F), non-MGMTmet (H), and MGMTmet (I).
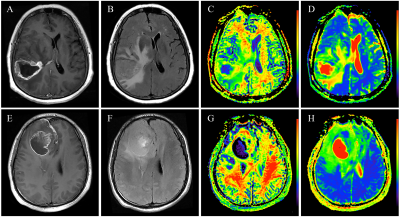
Figure 3 The high-grade astrocytoma showing ring enhancement in high- and low-risk patients. One high-risk patient (Radioscore=0.30; combination therapy; non-MGMTmet; OS=17.77 months) is illustrated in legends A-D. One low-risk patient (Radioscore=-0.204; combination therapy; MGMTmet; OS=38.73 months) is illustrated in legends E-H. Note: (A), (E) represent CE-T1WI images; (B), (F) represent T2FLAIR images; (C), (G) represent MK images; (D), (H) represent MD images.
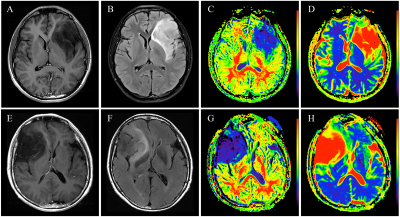
Figure 4 The low-grade astrocytoma without enhancement in high- and low-risk patients. One high-risk patient (Radioscore=0.121; surgery; non-MGMTmet; OS=21.67 months) is illustrated in legends A-D. One low-risk patient (Radioscore= -0.107; surgery; MGMTmet; OS=33.7 months) is illustrated in legends E-H. Note: (A), (E) represent CE-T1WI images; (B), (F) represent T2FLAIR images; (C), (G) represent MK images; (D), (H) represent MD images.
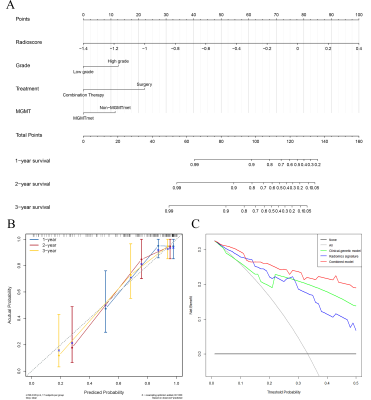
Figure 5 (A) Nomogram integrated radiomics signature, grade, treatment, and MGMT. (B) Calibration curves evaluated the consistency between predicted and actual survival probability. (C) Decision curve analysis of clinical-genetic model, radiomics signature and combined model.
DOI: https://doi.org/10.58530/2023/2884