2841
New Multiple Sclerosis Lesion Segmentation in Longitudinal FLAIR MR Images using Subtraction Image1Department of Biomedical engineering, Hankuk University of Foreign Studies, Yongin, Korea, Republic of, 2College of Medicine, The Catholic University of Korea, Seoul, Korea, Republic of, 3Department of Radiology, Seoul St. Mary's Hospital, Seoul, Korea, Republic of
Synopsis
Keywords: Multiple Sclerosis, Multiple Sclerosis, longitudinal MR, new lesion
Measurement of new lesions in longitudinal images is crucial in the evaluation of MS but is laborious and time-consuming. In this study, wWe investigate the effect of subtraction images between two time point FLAIR images on new MS lesion segmentation in longitudinal FLAIR images. The results showed that our method with combination of longitudinal FLAIR images and their subtraction images together helps to improve the accuracy of segmentation of new MS lesion.Introduction
The most common neuroinflammatory disease of the central nervous system, multiple sclerosis(MS), is marked by demyelination and persistent inflammation 1. Detecting new lesions in longitudinal FLAIR MRI scans is crucial in the diagnosis and management of MS patients since dissemination in time and space is one of the main clinical and imaging aspects of the disease 2. For diagnosis and research, an automated and trustworthy method would be convenient because detecting lesion changes manually requires a lot of time and is subject to intra- and inter-observer differences 3.Recently, numerous automated techniques to identify existing MS lesions and new MS lesions have been developed 4-6. However, there are still a number of difficulties mainly due to the fact that the volume of new lesions are small and are scattered throughout the central nervous system. On the other hand, Subtraction images between two timepoints show to improve the contrast between active lesions and the background because inactive lesions cancel each other out 7,8. In this study, we propose a framework for finding new multiple sclerosis lesions using deep learning models trained in parallel network architecture using two time points longitudinal dataset and their subtraction images.
Methods
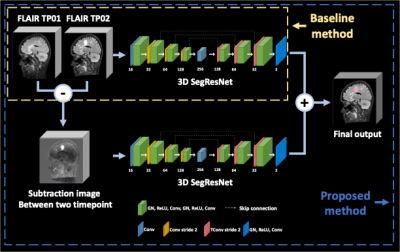
[Dataset and Preprocessing] In this study, the data of 100 patients from the MSSEG2 Challenge in MICCAI22 were utilized. A pair of 3D FLAIR images collected at different time points made up the dataset. One to three years after the first 3D FLAIR sequence, a second one was obtained, and masks of new MS lesions that were confirmed by 4 experts were provided. The FLAIR images were acquired from 15 different scanner models including both 1.5T and 3T scanners. For training and testing, we used the training (N=40) and testing (N=60) sets as they were divided by the MSSEG2 challenge. This dataset was provided after co-registration between the two timepoints. Also, to reduce the non-uniform bias field before training, we conducted the N4 bias field correction for preprocessing. Additionally, we used a brain mask from FMRIB Software Library(FSL v 0.6) and spinal cord from seed-based region growing for reduced false positives located outside the central nervous system.[Deep learning architecture] Our proposed method is summarized in Figure 1. The two parallel SegResNet 9 architectures were trained with different inputs. First network takes 2 channel image consist of two timepoint images of 96✕96✕96 used two timepoint FLAIR images, and second network takes 1 channel image of subtraction images between two timepoint images of 96✕96✕96. Then, the outputs of each model were averaged to produce the final output. We extracted random 96✕96✕96 patches, that preserved the ratio of samples with and without new MS lesion for training.To reduce overfitting and increase generality of the training model, we applied random transform of affine, intensity shifting, gaussian noise, gaussian smoothing, flipping, rotating. Networks were trained using AdamW optimizer with the initial learning rate of 0.003 and a weight decay of 0.00001 10. To update the network parameters, we used a combination of Cross-Entropy and Dice similarity coefficient loss function.
[Evaluation] We evaluate segmentation performance with the two input data combinations to test the impact of subtraction images on the segmentation of new MS lesions in Figure 1. For quantitative comparison, the F1 score, dice similarity coefficient, true-positive rate, and positive-predictive value were used to assess the performance of segmenting subjects with new MS lesions. The number and the volume of predicted lesions were calculated for subjects without new MS lesions.
Results
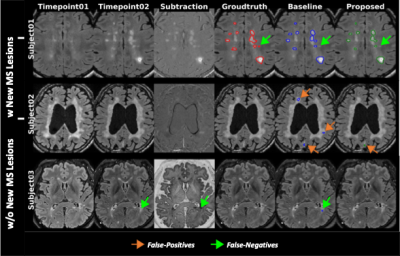
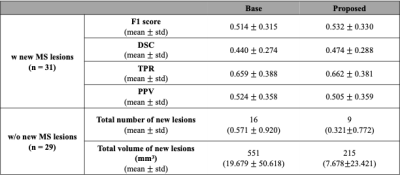
Figure 2 shows representative images of the result from two different input data combinations. The results showed that similar sensitivity for new MS lesions, but the proposed method markedly reduced false positive detections for subjects without new MS lesions. Table 1 summarizes the quantitative metrics calculated from the testing set for 60 subjects. For all cases, the proposed method shows higher scores in most quantitative metrics. When the proposed model with subtraction images was used, both datasets showed higher performance compared to the models without the improved subtraction images.Conclusion
We propose a detection method combining two time points longitudinal FLAIR images and their subtraction images that could improve the detection of new MS lesions. The proposed method improved both sensitivity and specificity when compared to the case where only longitudinal FLAIR images were used as input. This improvement may help to detect and measure new MS lesions in longitudinal FLAIR images in practice.Acknowledgements
This work was carried out in collaboration with The Observatoire Français de la Sclérose en Plaques (OFSEP), who is supported by a grant provided by the French State and handled by the“Agence Nationale de la Recherche,” within the framework of the “Investments for the Future” program, under the reference ANR-10-COHO-002, by the Eugène Devic EDMUS Foundation against multiple sclerosis and by the ARSEP Foundation. This work was supported by a grant (NRF-2020R1F1A1070517) of the Basic Science Research Program through the National Research Foundation of Korea.References
1. Gaetani, L.; Prosperini, L.; Mancini, A.; Eusebi, P.; Cerri, M. C.; Pozzilli, C.; Calabresi, P.; Sarchielli, P.; Di Filippo, M. 2017 revisions of McDonald criteria shorten the time to diagnosis of multiple sclerosis in clinically isolated syndromes. J. Neurol. 2018, 265, 2684-2687.
2. Egger, C.; Opfer, R.; Wang, C.; Kepp, T.; Sormani, M. P.; Spies, L.; Barnett, M.; Schippling, S. MRI FLAIR lesion segmentation in multiple sclerosis: does automated segmentation hold up with manual annotation? NeuroImage: Clinical 2017, 13, 264-270.
3. Ashton, E. A.; Takahashi, C.; Berg, M. J.; Goodman, A.; Totterman, S.; Ekholm, S. Accuracy and reproducibility of manual and semiautomated quantification of MS lesions by MRI. Journal of Magnetic Resonance Imaging: An Official Journal of the International Society for Magnetic Resonance in Medicine 2003, 17, 300-308.
4. Krüger, J.; Opfer, R.; Gessert, N.; Ostwaldt, A.; Manogaran, P.; Kitzler, H. H.; Schlaefer, A.; Schippling, S. Fully automated longitudinal segmentation of new or enlarged multiple sclerosis lesions using 3D convolutional neural networks. NeuroImage: Clinical 2020, 28, 102445.
5. McKinley, R.; Wepfer, R.; Grunder, L.; Aschwanden, F.; Fischer, T.; Friedli, C.; Muri, R.; Rummel, C.; Verma, R.; Weisstanner, C. Automatic detection of lesion load change in Multiple Sclerosis using convolutional neural networks with segmentation confidence. NeuroImage: Clinical 2020, 25, 102104.
6. Commowick, O.; Cervenansky, F.; Cotton, F.; Dojat, M. In In MSSEG-2 challenge proceedings: Multiple sclerosis new lesions segmentation challenge using a data management and processing infrastructure; MICCAI 2021-24th International Conference on Medical Image Computing and Computer Assisted Intervention; 2021; , pp 1-118.
7. Moraal, B.; Meier, D. S.; Poppe, P. A.; Geurts, J. J.; Vrenken, H.; Jonker, W. M.; Knol, D. L.; van Schijndel, R. A.; Pouwels, P. J.; Pohl, C. Subtraction MR images in a multiple sclerosis multicenter clinical trial setting. Radiology 2009, 250, 506.
8. Moraal, B.; Wattjes, M. P.; Geurts, J. J.; Knol, D. L.; van Schijndel, R. A.; Pouwels, P. J.; Vrenken, H.; Barkhof, F. Improved detection of active multiple sclerosis lesions: 3D subtraction imaging. Radiology 2010, 255, 154-163.
9. Myronenko, A. In In 3D MRI brain tumor segmentation using autoencoder regularization; International MICCAI Brainlesion Workshop; Springer: 2018; , pp 311-320.10. Loshchilov, I.; Hutter, F. Decoupled weight decay regularization. arXiv preprint arXiv:1711.05101 2017.
Figures