2797
Unsupervised Super-Resolution of Magnetic Resonance Images Using Deep Image Prior1Northwestern Polytechnical University, Xi'an, China, 2University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
Synopsis
Keywords: White Matter, Diffusion/other diffusion imaging techniques
The anatomical resolution of MRI is typically limited by acquisition time constraints. While deep learning networks have shown great potential for post-acquisition MRI resolution enhancement, their training typically relies on low-high resolution image pairs, which are not always available in practice. Here, we propose using deep image prior (DIP) for unsupervised MRI resolution enhancement with network training relying only on low-resolution images. Experimental results indicate that our method super-resolve MR images effectively with realistic details.
Purpose
Resolution increase in MRI is associated with prolonged scan time and reduced signal-to-noise ratio. While deep neural networks have shown remarkable performance in post-acquisition MRI super-resolution 1-3, they are typically trained supervised with high- and low-resolution image pairs. Here, we propose a DIP 4 framework to generate high-resolution MRI images with de novo training based on individual low-resolution images. Experimental results indicate that our method generates high-resolution MR images with realistic details and outperforms various competing methods.Methods
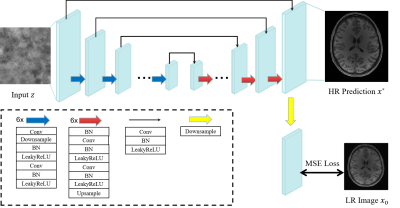
In our DIP framework, a high-resolution MR image $$$x^{*} \in \mathcal{R}^{H \times W}$$$ is predicted by a convolutional neural network $$$f_{\theta^{*}}(\cdot)$$$ with weights $$$\theta^{*}$$$ and random-noise input $$$z$$$: $$x^{*}=f_{\theta^{*}}(z)$$ Given a low-resolution image slice $$$x_{0} \in \mathcal{R}^{\frac{H}{t} \times \frac{W}{t}}$$$ downsampled by a factor $$$t$$$, the network weights are obtained via:$$\theta^{*}=\underset{\theta}{\operatorname{argmin}} E\left(f_{\theta}(z) , x_{0}\right)$$The energy function is: $$E\left(x, x_{0}\right)=\left\|d(x)-x_{0}\right\|^{2}$$ where $$$\|\cdot\|$$$ is the $$$\ell_2$$$ norm, $$$d(\cdot): \mathcal{R}^{t H \times t W} \rightarrow \mathcal{R}^{H \times W}$$$ is an operator that downsamples the high-resolution image $$$x=f_{\theta}(z)$$$ with factor $$$t$$$. Regularization is achieved with early stopping to avoid overfitting to the low-resolution image.The backbone of our network is a U-Net-like5 architecture (Fig. 1) with six skip-connections, six downsampling modules, and six upsampling modules. Downsampling of the output is performed by average pooling6 with factor $$$t$$$.
Results
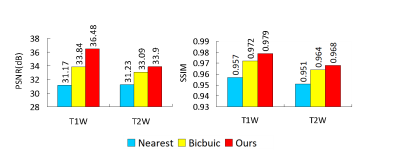
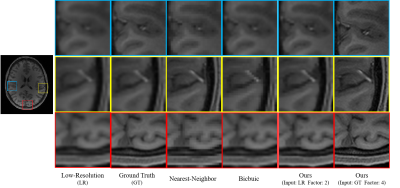
Dataset: We evaluated our method using T1- and T2-weighted MR images of five subjects from the human connectome project (HCP)7. We randomly selected slices from the MR images and then removed the non-brain background using a bounding box of size 256x288. We simulated low-resolution images by local averaging, as described in 6. Note that only low-resolution images are involved in training. High-resolution images are used as ground truth for evaluation.Experimental Results: We considered an undersampling factor of two and compared our method with bicubic and nearest-neighbor interpolation. The PSNR and SSIM results (Fig. 2) indicate that our method outperforms conventional interpolation methods, especially for T1W images. Visual results (Fig. 3) indicate that our method results in clearer anatomical details. Directly super-resolving the ground truth high-resolution images by a factor of four, our method further improves anatomical details (Fig. 3).
Conclusion
We have proposed a DIP-inspired deep learning framework for the super-resolution of MR images, without requiring high-resolution images for training. Method effectiveness was demonstrated qualitatively and quantitatively using the HCP data.Acknowledgements
Pew-Thian Yap was supported in part by the United States National Institutes of Health (NIH) under Grant EB008374 and Grant EB006733.References
[1] Chen, Yuhua, et al. "Brain MRI super resolution using 3D deep densely connected neural networks." 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018). IEEE, 2018.
[2] Masutani, Evan M., Naeim Bahrami, and Albert Hsiao. "Deep learning single-frame and multiframe super-resolution for cardiac MRI." Radiology 295.3 (2020): 552.
[3] de Leeuw den Bouter, M. L., et al. "Deep learning-based single image super-resolution for low-field MR brain images." Scientific Reports 12.1 (2022): 1-10.
[4] Ulyanov, Dmitry, Andrea Vedaldi, and Victor Lempitsky. "Deep image prior." Proceedings of the IEEE conference on computer vision and pattern recognition. 2018.
[5] Manjón, José V., et al. "Non-local MRI upsampling." Medical image analysis 14.6 (2010): 784-792.
[6] Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. "U-Net: Convolutional networks for biomedical image segmentation." International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015.
[7] Van Essen, David C., et al. "The WU-Minn human connectome project: an overview." Neuroimage 80 (2013): 62-79.