2776
Regional Achilles Tendon Segmentation for Quantitative Magnetic Resonance Imaging at 1.5 Tesla1Department of Cybernetics and Biomedical Engineering, VSB Technical University of Ostrava, Ostrava, Czech Republic, 2Department of Biomedical Imaging and Image-guided Therapy, High Field MR Centre, Medical University of Vienna, Vienna, Austria, 3Department of Human Movement Studies, Human Motion Diagnostic Center, University of Ostrava, Ostrava, Czech Republic, 4Department of Radiology, Vitkovice Hospital, Ostrava, Czech Republic, 5Department of Imaging Methods, Faculty of Medicine, University of Ostrava, Ostrava, Czech Republic, 6Department of Radiology, University Hospital Ostrava, Ostrava, Czech Republic, 7Institute of Measurement Science, Slovak Academy of Sciences, Bratislava, Slovakia, 8CD Laboratory fo MR Imaging Biomarkers (BIOMAK), Vienna, Austria, 9Austrian Cluster for Tissue Regeneration, Ludwig Boltzmann Institute for Experimental and Clinical Traumatology, Vienna, Austria, 10Institute for Clinical Molecular MRI in the Musculoskeletal System, Karl Landsteiner Society, Vienna, Austria
Synopsis
Keywords: Tendon/Ligament, Machine Learning/Artificial Intelligence
Changes in the Achilles tendon composition are associated with an increased risk of tendinopathy which is common in middle-aged overweight patients and is one of the most common sports injuries. However, measuring and quantifying such properties is a challenging task. The purpose of this paper is to introduce an end-to-end pipeline for segmenting Achilles tendon using deep convolutional neural networks and automatic segmentation into three segments. Our model shows promising results outperforming state-of-the-art approaches (Dice 90.6% and Jaccard 84.0%). This is one of the key steps for short and long T2* value analysis from 1.5T data.Introduction
The incidence of Achilles tendinopathy has increased over the past few decades due to an increase in recreational and competitive sports participation as well as increased workloads1,2. The use of magnetic resonance imaging (MRI) can provide an understanding of the internal morphology of tendons and their external anatomy as well as quantitative characteristics of tissues, such as T2 and T2* maps. These measurable features could be further used for predicting or monitoring the progression of tendinopathy3–5. The process of measuring quantitative characteristics of tissues is laborious and time-consuming, requires a great deal of user interaction. Often, these interactions introduce a intraobserver and interobserver variability which directly affects reproducibility. Therefore, there is a lot of effort put into creating a reliable and less biased approach based on automatic segmentation6–8. This study aims to develop a fully automated Achilles tendon segmentation pipeline using Deep Convolutional Neural Networks (CNN).Methods
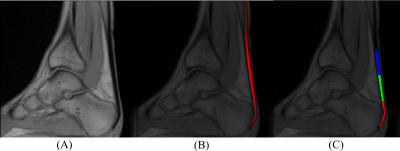
The MR data used in this study were acquired using a Siemens MAGNETOM Sempra 1.5T scanner with a 16-channel head coil. A total of 400 unpaired ankle scans of healthy volunteers were acquired using two T2* sequences in the sagittal plane with the following parameters: TR/TE1 = 485/3.78, 10.77, 17.15, 23.52, 29.89 ms, TR/TE2 = 485/7.28, 14.28, 21.28, 28.28, 35.28 ms, resolution = 0.6x0.6x3 mm, FA = 60° with TA = 3:20 minutes. All images were scored by a radiologist using the VIMATS scoring system9. Data annotations were performed by a trained image analyst (D.V., with 3 years of experience) using in-house written MATLAB scripts (version 2020b, MathWorks Inc., Natick, MA, USA). All segmentations were reviewed by a radiologist with 10 years' experience (M.G.). The 2D U-Net10 was used and all data has been augmented to avoid overfitting. A total of 320 patients were used as training data and 80 patients as test data. A quantitative evaluation of model performance was conducted using Dice similarity coefficient and Jaccard similarity index. Finally, the Achilles tendon was automatically divided into three portions: the insertion, the mid portion, and the junction with the muscle tendon.Results
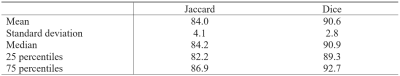
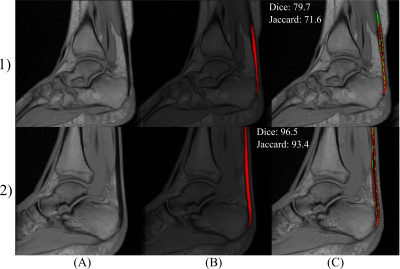
The VIMATS grading performed by the radiologist shows 97 ± 4.9%, indicating that the testing dataset consists mostly of physiological subjects. Table 1 shows the results of a deep CNN model performance based on objective metrics (Dice and Jaccard). Figure 1 shows the examples of the best and the worst segmentation results according to the Dice similarity coefficient. Then, all the segmentations were automatically separated into three portions using custom Python code (version 3.8.), see Figure 2.Discussion and Conclusion
This study presents an end-to-end pipeline for segmenting Achilles tendon from 1.5T T2* images based on a deep CNN approach. Using this approach, Achilles tendon is automatically segmented and divided into three clinically relevant portions. Our model achieved promising results (Dice 90.6 ± 2.8 % and Jaccard 84.0 ± 4.1 %) outperforming the state-of-the-art models. This approach is one of the crucial steps for automatic quantitative evaluation of Achilles tendon properties. Despite the promising results and advantages, our study has the limitation that the 3D segmentation could not be performed as this study only involved data from 1.5T scanner. The future research aims to investigate the quantitative means of evaluating short and long T2* values of the Achilles tendon in different tensile regions from 1.5T data. This holds significant promise for fully automated quantification of Achilles tendon from images acquired in lower fields. Moreover, we intend to retrain the presented model on data acquired from 3T and 7T data. In addition, other deep CNN models will be compared to improve the accuracy and performance of automatic segmentation.Acknowledgements
This study was supported by the Austrian Science Fund, KLI 917, by the Ministry of Education of the Czech Republic under Project SP2021/32 and co-funded by European union and Ministry of Education, Youth and Sports of the Czech Republic, grant number CZ.02.1.01/0.0/0.0/16_019/0000798 Program 4 Healthy Aging in Industrial Environment.References
1. Longo UG, Ronga M, Maffulli N. Achilles Tendinopathy. Sports Medicine and Arthroscopy Review. 2018;26(1):16-30. doi:10.1097/JSA.0000000000000185
2. Vadalà A, Lanzetti RM, Ciompi A, Rossi C, Lupariello D, Ferretti A. Functional evaluation of professional athletes treated with a mini-open technique for achilles tendon rupture. Muscles Ligaments Tendons J. 2014;4(2):177-181.
3. Malmgaard‐Clausen NM, Tran P, Svensson RB, et al. Magnetic Resonance T 2 * Is Increased in Patients With Early‐Stage Achilles and Patellar Tendinopathy. Magnetic Resonance Imaging. 2021;54(3):832-839. doi:10.1002/jmri.27600
4. Juras V, Zbyn S, Pressl C, et al. Regional variations of T 2 * in healthy and pathologic achilles tendon in vivo at 7 Tesla: Preliminary results: T 2 * in Healthy and Pathologic AT at 7 T. Magn Reson Med. 2012;68(5):1607-1613. doi:10.1002/mrm.24136
5. Devaprakash D, Obst SJ, Lloyd DG, et al. The Free Achilles Tendon Is Shorter, Stiffer, Has Larger Cross-Sectional Area and Longer T2* Relaxation Time in Trained Middle-Distance Runners Compared to Healthy Controls. Front Physiol. 2020;11:965. doi:10.3389/fphys.2020.00965
6. Regulski PA, Zielinski J. Multi-Step Segmentation Algorithm for Quantitative Magnetic Resonance Imaging T2 Mapping of Ruptured Achilles Tendons. IEEE Access. 2020;8:199995-200004. doi:10.1109/ACCESS.2020.3035549
7. Alzyadat T, Praet S, Chetty G, et al. Automatic Segmentation of Achilles Tendon Tissues Using Deep Convolutional Neural Network. In: Liu M, Yan P, Lian C, Cao X, eds. Machine Learning in Medical Imaging. Vol 12436. Lecture Notes in Computer Science. Cham: Springer International Publishing; 2020:444-454. doi:10.1007/978-3-030-59861-7_45
8. Juras V, Apprich S, Szomolanyi P, Bieri O, Deligianni X, Trattnig S. Bi-exponential T2* analysis of healthy and diseased Achilles tendons: an in vivo preliminary magnetic resonance study and correlation with clinical score. Eur Radiol. 2013;23(10):2814-2822. doi:10.1007/s00330-013-2897-8
9. Apprich S, Nia A, Schreiner MM, Friedrich K, Windhager R, Trattnig S. The Vienna morphological Achilles tendon score—VIMATS: Description, reproducibility and initial clinical results. Wien Klin Wochenschr. 2021;133(11-12):560-567. doi:10.1007/s00508-021-01863-6
10. Ronneberger O, Fischer P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, eds. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Vol 9351. Lecture Notes in Computer Science. Cham: Springer International Publishing; 2015:234-241. doi:10.1007/978-3-319-24574-4_28
Figures