2746
Weakly supervised learning improves vascular territorial mapping of random vessel-encoded ASL1Radiology, Northwestern University, Chicago, IL, United States, 2Neurology, University of Southern California, Los Angeles, CA, United States
Synopsis
Keywords: Arterial spin labelling, Perfusion
This study proposed a weakly supervised learning algorithm for vascular territorial mapping with rVE-ASL. The territory maps generated by the proposed deep learning (DL) method was compared with the territories from the conventional rVE-ASL method by visual inspection and F1 score. Our initial results showed that the DL method outperformed the conventional rVE-ASL method in the vascular territory mapping with improved detection of VA territory. The DL method also provided reliable vascular territorial maps with reduced numbers of encodings, significantly reducing rVE-ASL scan time. These findings suggest that DL could be an effective approach for vascular territorial mapping of rVE-ASL.Introduction
Characterization of cerebral blood flow from individual arteries is important in clinical applications. Vessel-encoded ASL (VE-ASL) enables the vascular territorial maps from individual feeding arteries but with the need of prior knowledge of the positions of feeding arteries1. To get rid of the limitation, random vessel-encoded ASL (rVE-ASL)2 has been developed, which can generate vascular territories and corresponding feeding arteries simultaneously by modulating labeling efficiency patterns in the labeling plane through a number of random encoding steps. There are several limitations in rVE-ASL. First, multiple encoding steps are required in order to achieve reliable territorial mapping, which results in relatively long scan time. Second, the perfusion territory of the vertebral artery (VA) often fails to be detected using the current rVE-ASL processing method due to the low labeling efficiency of VA. Recently, deep learning algorithms3 have been developed to cluster feature vectors into different categories. In this study, we aim to develop a semi-supervised deep clustering algorithm to improve the sensitivity of vascular territory detection in rVE-ASL.Methods
Imaging protocolAll MRI experiments were conducted on a Siemens 3T scanner. rVE-ASL sequence with background suppressed (BS) 3D Gradient And Spin Echo (GRASE) was performed on seven healthy volunteers with the following parameters: FOV= 220×220 mm2, matrix size=64×64, TE/TR=22/3500ms, 16 slices with slice thickness of 6mm, 60 pairs of encoding steps with random orientation, phase and wavelength were acquired with two additional pairs of global label/control. VE-ASL scan was also performed on each participant with closely matched imaging parameters. VE-ASL images were decoded into vascular territory maps of left ICA, right ICA, and VA.
Network architecture
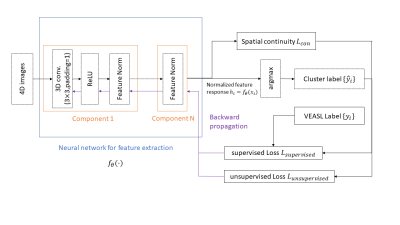
The neural network architecture is shown in Figure 1. 4D rVE-ASL data are the input of the neural network. The rVE-ASL data from all subjects were used for deep learning. The neural network outputs a normalized feature response $$$h_i = f_{\theta}(x_i)$$$ . Predicted voxel-wise vascular territory index $$${\hat{y}_i}$$$ is computed by applying argmax function on normalized feature response.
Training method
We proposed a semi-supervised learning for feature learning and prediction of vascular territory index, which combines supervised learning from VE-ASL generated vascular territory indices as initialization and unsupervised feature learning from rVE-ASL validation data. Based on the visual inspection, three VE-ASL datasets with reliable territorial maps (i.e., all three territories were successfully detected) were chosen as ground truth for the supervised training. The remaining 4 subjects’ data were used for unsupervised training and validation.
Loss function
The feature similarity loss $$$ L_{sim}(f_{\theta}(x_i),\hat{y_i}) $$$ is computed by using cross entropy function to maximize the mutual information between normalized feature response and target territory index.
$$L_{sim}(f_{\theta}(x_i,\hat{y_i})) = \sum_{i=1}^{N}\sum_{j=1}^SL_{cross entropy}(f_{\theta}(x_{i,j},\hat{y_{i,j}}))$$
where i is the subject index, and j is the spatial index for each voxel. Here the cross-entropy loss is defined by
$$L_{crossentropy}(x_i,y_i) = - \sum_{c=1}^Cw_clog\frac{exp(x_{i,c})}{\sum_{j=1}^Cexp(x_{i,j})}y_{i,c}$$
y is vascular territory index; $$$x_{i,j}$$$ is input data and j is the class index. $$$w_c$$$ is the class weight. Here we set weight of vertebral artery index as 2 (others are 1) to enhance neural network training on vertebral artery. The spatial continuity $$$L_{con}$$$ of $$$h_i=f_{\theta}(x_i)$$$ is constrained to ensure feature similarity among neighboring voxels.
$$L_{con}(h_i) = \sum_{ix=1}^{W-1} \sum_{iy=1}^{H-1} \sum_{iz=1}^{D-1} \|h_{ix+1,iy,iz} - h_{ix,iy,iz} \|_1+ \|h_{ix,iy+1,iz} - h_{ix,iy,iz} \|_1+ \|h_{ix,iy,iz+1} - h_{ix,iy,iz} \|_1$$
The loss for supervised learning is
$$ loss_{supervised} = L_{sim}(f_{\theta}(x_i),y_i) +\lambda L_{sim}(f_{\theta}(x_i),\hat{y_i})+\mu L_{con}(f_{\theta}(x_i))$$
The loss for unsupervised learning is
$$loss_{unsuperivsed} = \lambda L_{sim}(f_{\theta}(x_i),\hat{y_i})+\mu L_{con}(f_{\theta}(x_i))$$
Results
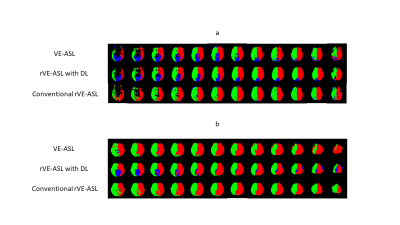
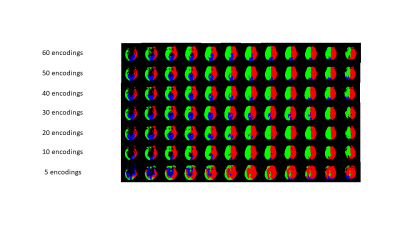
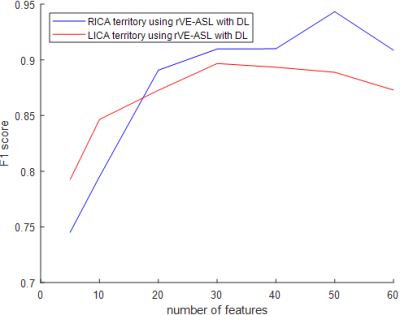
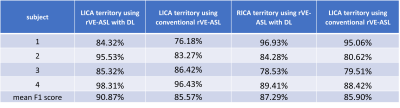
Figure 2a shows an example of the vascular territory maps generated by VE-ASL, rVE-ASL with proposed DL, and conventional rVE-ASL processing method2, respectively. VE-ASL and rVE-ASL territory maps with DL matched closely with each other, both of which successfully detected all three territories. In contrast, conventional rVE-ASL method failed to detect VA territory. Figure 2b shows another case, in which both VE-ASL and conventional rVE-ASL failed to detect VA territory, whereas VA territory was successfully retrieved by using DL method, suggesting DL improves the territorial detection including VA territory. The performance of the rVE-ASL with the DL and conventional methods were compared quantitatively using F1 scores from the validation data (Table 1). DL outperformed the conventional rVE-ASL in LICA and RICA territorial mapping. No F1 scores were calculated for VA territory, as no VA territories were detected by the conventional rVE-ASL in all four subjects, and VE-ASL only detected VA territory from one of them. However, using DL, VA territory was successfully retrieved from rVE-ASL in three subjects. Figure 3 shows the perfusion territorial maps with different numbers of encodings using DL. Reliable perfusion territories were preserved until the number of encodings reduced to 30. Quantitative evaluation shown in Figure 4 further confirmed the findings in Figure 3.Discussion & Conclusion
In this study, we proposed a deep learning clustering algorithm to compute perfusion territory maps from rVE-ASL. Our initial results suggest that deep learning provides more reliable perfusion territory mapping with improved detection of VA territory, compared to the conventional rVE-ASL method. Using deep learning, the total encoding steps can significantly decrease while maintaining good performance on the detection of perfusion territories. Future work with larger sample size and further optimization is needed to improve the robustness of the proposed DL model. Validation on a relatively heterogeneous cohort will be carried out.Acknowledgements
This work is supported by grants of NIH R01NS118019, RF1AG072490, and BrightFocus Foundation A20201411S.References
1. Wong EC. Vessel‐encoded arterial spin‐labeling using pseudocontinuous tagging. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine. 2007 Dec;58(6):1086-91.
2. Wong EC, Guo J. Blind detection of vascular sources and territories using random vessel encoded arterial spin labeling. Magnetic Resonance Materials in Physics, Biology and Medicine. 2012 Apr;25(2):95-101.
3. Kim W, Kanezaki A, Tanaka M. Unsupervised learning of image segmentation based on differentiable feature clustering. IEEE Transactions on Image Processing. 2020 Jul 28;29:8055-68.
4. Kansagra AP, Wong EC. Improved estimation of cerebral artery branch territories using cluster-based segmentation of vessel-encoded pseudocontinuous ASL data. In Proceedings of the 20th Annual Meeting ISMRM Melbourne, Australia 2012 (Vol. 583).
Figures