2745
Multi-2D ASL-MRA and super-resolution convolutional neural network for improved intracranial peripheral arteries visualization
Yuriko Suzuki1, Ioannis Koktzoglou2,3, Peter Jezzard1, and Thomas Okell1
1Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom, 2Department of Radiology, NorthShore University HealthSystem, Evanston, IL, United States, 3Pritzker School of Medicine, University of Chicago, Chicago, IL, United States
1Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom, 2Department of Radiology, NorthShore University HealthSystem, Evanston, IL, United States, 3Pritzker School of Medicine, University of Chicago, Chicago, IL, United States
Synopsis
Keywords: Arterial spin labelling, Blood vessels, MRA
The non-invasive nature of Arterial Spin Labeling (ASL) technique makes ASL-based intracranial dynamic MR angiography (MRA) a potential alternative to diagnostic X-ray digital subtraction angiography. In elderly and diseased patients with slower blood flow, however, the vessel visualization of distal peripheral arteries tends to be poor, as the repeatedly applied RF excitation pulses decrease ASL blood signal rapidly. In this study, we address such a limitation by using multiple 2D slice acquisition to reduce the saturation of arterial blood signal. Additionally, to avoid losing vessel conspicuity and sharpness with 2D slice acquisition, we apply a super-resolution convolutional neural network method.Introduction
The non-invasive nature of the Arterial Spin Labeling (ASL) technique, as well as the ability to achieve vessel-selective visualization, makes ASL-based dynamic MR angiography (MRA) a potential alternative to X-ray digital subtraction angiography (DSA) for diagnosis. However, to make ASL-MRA as effective as conventional X-ray-DSA, there is an important limitation to be addressed; vessel visualization tends to be poor when blood flow is slow, which is common in elderly patients and in patients with arterial disease. This problem is largely attributable to 3D volume acquisition, which is typically used in brain MRA to cover a large volume of brain with high spatial resolution. The repeated application of RF excitation pulses to the whole volume for 3D imaging causes a rapid decrease of ASL arterial blood signal while it flows more distally into the brain.This project aims to develop a novel framework to improve the visualization of peripheral arteries in elderly patients by using the following three approaches: (i) the exposure of flowing arterial blood to the excitation pulses is reduced by using multiple 2D (M2D) slice acquisition. With conventional MRI scanners, however, it is difficult to achieve the thin slice thickness required for brain MRA. Therefore, (ii) the spatial resolution in the slice-direction is improved by applying a super-resolution convolutional neural network (SR-CNN) method1. Additionally, (iii) the SR-CNN is trained by using publicly available time-of-flight (TOF) MRA datasets, to avoid the need to acquire a large number of ASL-MRA images for network training.
Methods
Three subjects were scanned on a Siemens 3T Prisma under a technical development protocol agreed by local ethics and institutional committees. As Figure-1 indicates, M2D dynamic ASL-MRA was acquired with pseudo-continuous ASL (PCASL) labeling and a 2D FLASH readout. After each PCASL labeling (500ms), a single 2D slice was excited (3mm thickness, flip angle = 26°) and the readout was repeated 6 times in a Look-Locker readout manner. In the subsequent labelings, multiple 2D slices were scanned one by one with 50% (1.5mm) overlap with the previous excitation. With a total of 60 slices, the scan time was 6:59. For comparison, dynamic ASL-MRA with a 3D FLASH sequence was also acquired with the same imaging parameters (except 1.5mm slice thickness and flip angle = 10°), and the scan time was 6:33.Our SR-CNN consisted of seven 3D convolutions combined with rectified linear unit activation (except the last layer) and 64 filters1 (Figure-2). The training was performed by using forty 3T TOF-MRA volumes from the IXI Dataset (nearly 600 MR images were collected from normal, healthy subjects: https://brain-development.org/ixi-dataset/). Low-resolution (LR) data were generated by applying a Fourier transform to the original (high-resolution: HR) data along the slice-direction, zeroing the outermost 75% of k-space, and inverse Fourier transforming it back to the image domain. Subtraction is performed between HR and LR images to depict high-frequency information as we have low-frequencies from the LR image (Figure 2). Sets of the subtracted and LR images were divided into 32x32x32 voxel blocks; 500 blocks from each TOF volume were chosen from high signal intensity, aiming to extract the highest level of meaningful features. Another 200 blocks were also chosen randomly from the TOF field-of-view to condition the SR-CNN to reconstruct non-arterial background static tissue. The trained network was applied to additional ten TOF-MRA LR images from IXI Dataset for testing, as well as our M2D ASL-MRA images, to generate super-resolution images.
Results and Discussion
Figure-3 shows an example set of the test TOF-MRA images: (a) ground truth high-resolution, (b) low-resolution, and (c) super-resolution. The mean values of the structural similarity index (SSIM) indicate that the anatomical agreement as compared to the ground truth was significantly improved with application of the SR-CNN. Additionally, as shown in Figure-4, even though the SR-CNN was trained with TOF-MRA images, the SR-CNN successfully improved the vessel conspicuity and sharpness in M2D dynamic ASL-MRA as compared to (a) a standard temporal MIP (tMIP) and (b) a bicubically interpolated tMIP. Finally, Figure-5 shows a comparison between the conventional 3D dynamic ASL-MRA and M2D dynamic ASL-MRA images from the same subject, indicating that the visualization of peripheral arteries shown in later phases is improved in the M2D acquisition, without a loss in vessel conspicuity and sharpness. With the current sequence design, however, the scan time would be rather long to cover the entire brain; with 91.5mm coverage, the scan time was 6:59. In this study, the application of the SR-CNN to 3mm thickness images without overlap did not result in satisfactory image quality (data not shown). In future work, acceleration techniques will be attempted to shorten the scan time.Conclusion
In this study, we have shown that M2D ASL-MRA in conjunction with a super-resolution convolutional neural network was able to visualize more peripheral arteries with higher signal intensity than a conventional 3D ASL-MRA without losing the vessel conspicuity and sharpness. Furthermore, we found it beneficial that the SR-CNN applied to ASL-MRA data in the present study could be trained using TOF-MRA data, as the latter is more readily available and is of higher signal-to-noise ratio and spatial resolution.Acknowledgements
This project was supported by the Royal Academy of Engineering under the Research Fellowship scheme (RF\201920\19\236). The Wellcome Centre for Integrative Neuroimaging is supported by core funding from the Wellcome Trust (203139/Z/16/Z). PJ thanks the Dunhill Medical Trust and the Oxford NIHR Biomedical Research Centre for support. TO was supported by a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (Grant Number 220204/Z/20/Z). IK was supported by a grant from the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health (Award Number R01EB027475).References
1. Koktzoglou, I., Huang, R., Ankenbrandt, W. J., Walker, M. T. & Edelman, R. R. Super-resolution head and neck MRA using deep machine learning. Magn. Reson. Med. 86, 335–345 (2021).Figures
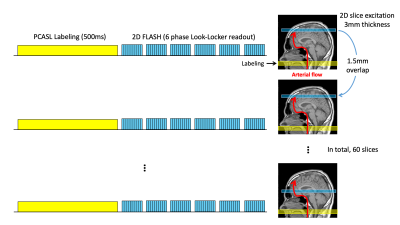
Figure 1: Schematic diagram of M2D dynamic ASL-MRA sequence, which consists of pseudo-continuous ASL (PCASL) labeling module and a 2D FLASH readout. After each PCASL labeling (500ms), a single 2D slice was excited (3mm thickness, flip angle = 26°) and the readout was repeated 6 times in a Look-Locker readout manner. In the subsequent labelings, multiple 2D slices were scanned one by one with 50% (1.5mm) overlap with the previous excitation. With a total of 60 slices, the scan time was 6:59.
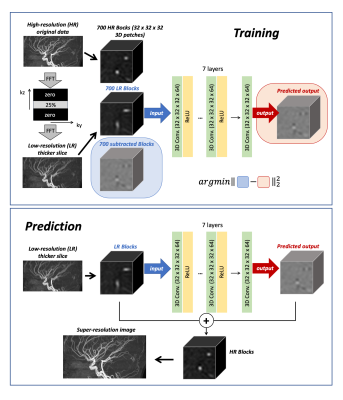
Figure 2: Schematic diagrams of the training and prediction processes using the super-resolution convolutional neural network (SR-CNN), which consists of seven 3D convolutions combined with rectified linear unit activation (except the last layer) and 64 filters.
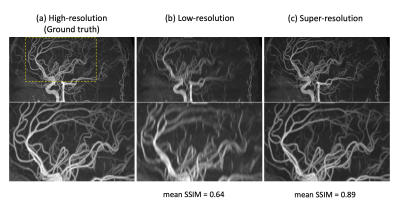
Figure 3: An example set of the test TOF-MRA images: (a) ground truth high-resolution, (b) low-resolution, and (c) super-resolution, as well as the mean values of the structural similarity index (SSIM) as compared to the ground truth.
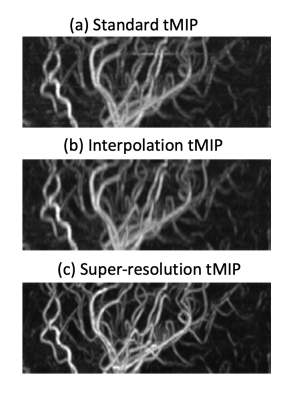
Figure 4: The temporal MIP (tMIP) of our M2D dynamic ASL-MRA. Even though the SR-CNN was trained with TOF-MRA images, (c) a super-resolution tMIP shows a notable improvement in the vessel conspicuity and sharpness as compared to (a) a standard tMIP and (b) a bicubically interpolated tMIP.
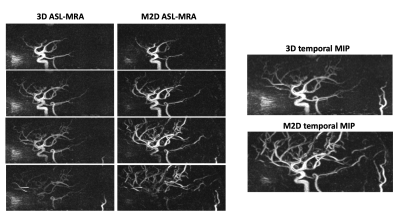
Figure 5: A comparison between the conventional 3D dynamic ASL-MRA and M2D dynamic ASL-MRA images from the same subject, indicating that the visualization of peripheral arteries shown in later phases is improved in the M2D acquisition, without a loss in vessel conspicuity and sharpness.
DOI: https://doi.org/10.58530/2023/2745