2738
Realistic Digital Reference Object (DRO) toolkit for quantitative breast Ultra-Fast Dynamic Contrast-Enhanced (UF-DCE) MRI1Vilcek Institute of Graduate Biomedical Science, NYU School of Medicine, New York, NY, United States, 2Center for Biomedical Imaging, Radiology, NYU School of Medicine, New York, NY, United States, 3Center for Advanced Imaging Innovation and Research, Radiology, NYU School of Medicine, New York, NY, United States, 4Radiology, Weill Cornell Medical College, New York, NY, United States, 5Biomedical Engineering, Friedrich-Alexander-Universitat Erlangen-Nurnberg, Erlangen, Germany
Synopsis
Keywords: Breast, DSC & DCE Perfusion
Reconstruction of highly accelerated dynamic images is challenging and requires rigorous validation of reconstruction methods. We proposed a digital reference object (DRO) toolkit that provides realistic morphology and contrast dynamics in breast cancer. We acquired various base images containing real breast cancer lesions and simulated realistic contrast dynamics using the estimated kinetic parameters. Our toolkit provides a large number of reference objects with wide ranges of the dynamic images, kinetic parameters, segmentation masks, and k-space data. These realistic DROs with known ground-truth values can be used for different studies, including validation of reconstruction and training deep neural network.Purpose
Ultra-fast Dynamic contrast-enhanced (UF-DCE) MRI aims to capture the early uptake of contrast dynamics in breast cancer patients, allowing the reduction of scan time from 10 min to less than 5 min (1). Quantitative analysis of UF-DCE-MRI requires a high temporal resolution (5~8s), while maintaining the relatively high spatial quality image, posing great challenges in image reconstruction. Although there are several advanced reconstruction schemes, validation of these suggested reconstruction is essential for the accurate analysis. Digital Reference Object (DRO) has been used for this purpose; however the current framework is limited to the random insertion of simple geometry (2) or limited shapes of simulated lesions (3). In this study, we aim to develop more realistic DRO toolkit for breast cancer studies.Methods
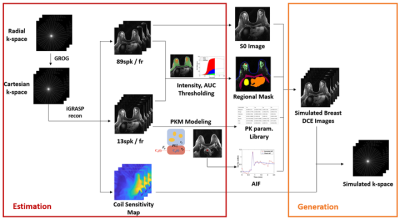
UF-DCE-MRI studyOur study recruited a total of 55 women who have either malignant (n=25, ages: 30-75 y/o) or benign (n=30, ages: 25-68 y/o) lesions. Each patient underwent UF-DCE-MRI exam, which was scanned at 3.0T with a radial stack-of-star 3D spoiled GRE sequence. The total scan time was 2.5min, while the contrast agent (gadobutrol, 0.1mM/kg body weight) was injected at 1 min into the scan. The scan parameters include TE/TR = 1.8/4.87ms, flip-angle=10 degrees, matrix size of 320x320x83 and the resolution of 1x1x1.1mm. The lesions were identified and segmented by a breast radiologist. For the image reconstruction, iterative GRASP reconstruction (4) was used to reconstruct dynamic images with 2 different settings: 89 spokes-per-frame for high spatial quality image and 13 spokes-per-frame for a high temporal resolution(6s).
Estimation Phase
We obtained the pre-contrast image from the high spatial quality image and used it for the baseline image. The high temporal quality image was used for segmentation and the pharmacokinetic model (PKM) analysis. The segmentation task was initiated by drawing a rough ROI manually around the targeted tissue, and further segmented by thresholding the AUC of early uptake up to 1.5min. We repeated the segmentation task for following regions: fibroglandular tissue, tumor lesions (malignant / benign), pectoral muscle, skin, liver, and heart. Then we performed bootstrapping to sample signals in each region and performed PKM analysis with two most widely used models in breast cancer study: the generalized kinetic model (GKM) and the two-compartment exchange model (TCM). The arterial input function (AIF) was acquired from the aorta using the ROCKETSHIP toolkit (5). The median values of estimated kinetic parameters for each region were collected from each case to form the range for simulation.
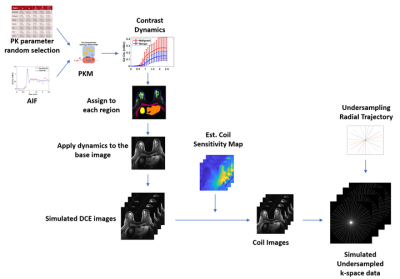
Generation Phase
From the estimated kinetic parameter range, we randomly selected kinetic parameters for each region. The selected parameters were added with 20% variations and paired with the selected AIF to simulate the contrast dynamics. Each region of contrast dynamics is then assigned to the baseline image to generate simulated DCE images, which are then combined with the estimated coil-sensitivity maps and the under-sampled radial trajectory to generate the simulated under-sampled k-space data.
Reconstruction Assessment
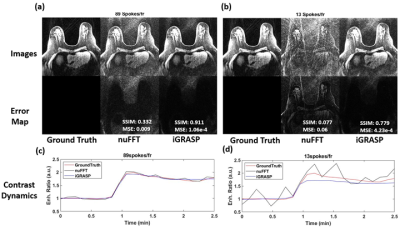
We provide an example application of our developed toolkit for assessment of reconstruction. We simulated a breast DCE image with a malignant lesion. We generated the k-space data at 2 different under-sampling rates: 89 and 13 spokes-per-frame. Then we employed 2 reconstruction schemes: first, we simply re-grid the radial data into the cartesian grid and reconstruct the image (nuFFT) and second, we performed iterative GRASP reconstruction (iGRASP) (4). To assess spatial quality, we measured the mean-squared error (MSE) and the structural similarity index measure (SSIM). For the temporal quality assessment, we performed the PKM analysis.
Result
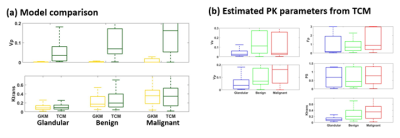
PKM analysis for DRO developmentThe Ktrans estimates from both GKM and TCM were not significantly different (p<0.05). However, we observed a consistent under-estimation of vp with GKM in all regions, as shown in Figure 3(a). This could be due to the lack of consideration for the vascular transport resulting in the severe underestimation of vp. Hence, we selected TCM for the toolkit. The Fp and PS estimation from TCM exhibited high variability, possibly due to the short scan time. However, the calculated Ktrans exhibited reduced variability with the physiologically reasonable range of values, as depicted in Figure 3(b). Therefore, we randomly simulated Fp and PS from the range, while constraining Ktrans.
Reconstruction Assessment
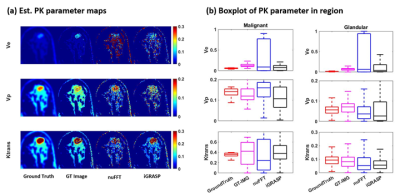
Figure 4 shows the spatial assessment of 2 different reconstruction scheme. When large number of spokes are used, both reconstruction methods provide similar images as the ground-truth. When the acquisition is accelerated, nuFFT images show the increased levels of streak artifact, while the iGRASP images suppress streak artifact and produce superior quality image as compared to nuFFT. However, iGRASP tends to underestimate the contrast dynamics, as shown in Figure 4(d). Figure 5 shows the temporal fidelity assessment via PKM analysis. The iGRASP images show accurate Ktrans estimation, but exhibits underestimation of vp, likely from the underestimation of contrast dynamics.
Discussion & Conclusion
We developed a realistic DRO for quantitative breast UF-DCE MRI. Our proposed toolkit, which is available as open source, can be used as a testbed for image reconstructions or for generating training data to train a machine learning network. Our future work includes the expansion of this framework in 3D and the simulation with different scan length.Acknowledgements
R01CA160620
R01CA219964
UH3CA228699
References
1. Kataoka M, Honda M, Ohashi A, Yamaguchi K, Mori N, Goto M, et al. Ultrafast Dynamic Contrast-enhanced MRI of the Breast: How Is It Used? Magnetic Resonance in Medical Sciences. 2022;21(1):83-94.
2. Wang PN, Velikina JV, Strigel RM, Henze Bancroft LC, Samsonov AA, Cashen TA, et al. Comparison of data‐driven and general temporal constraints on compressed sensing for breast DCE MRI. Magnetic resonance in medicine. 2021;85(6):3071-84.
3. Henze Bancroft L, Holmes J, Bosca-Harasim R, Johnson J, Wang P, Korosec F, et al. An Anthropomorphic Digital Reference Object (DRO) for Simulation and Analysis of Breast DCE MRI Techniques. Tomography. 2022;8(2):1005-23.
4. Feng L, Grimm R, Block KT, Chandarana H, Kim S, Xu J, et al. Golden‐angle radial sparse parallel MRI: combination of compressed sensing, parallel imaging, and golden‐angle radial sampling for fast and flexible dynamic volumetric MRI. Magnetic resonance in medicine. 2014;72(3):707-17.
5. Barnes SR, Ng TS, Santa-Maria N, Montagne A, Zlokovic BV, Jacobs RE. ROCKETSHIP: a flexible and modular software tool for the planning, processing and analysis of dynamic MRI studies. BMC medical imaging. 2015;15(1):1-20.
Figures