2708
Statistical evaluation of complexity tests for fMRI timeseries data1USC Mark and Mary Stevens Neuroimaging and Informatics Institute, Keck School of Medicine at USC, Los Angeles, CA, United States
Synopsis
Keywords: Software Tools, fMRI (resting state), Complexity, Higuchi Fractal Dimension, Hurst Exponent, Lempel-Ziv Complexity, Approximate Entropy, Multiscale Sample Entropy, Fuzzy Entropy, Permutation Entropy
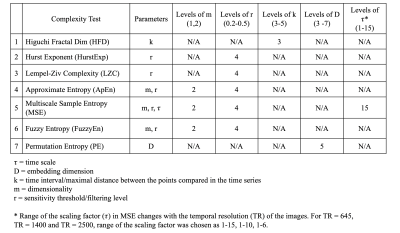
Complexity measures of rs-fMRI signals based on non-linear timeseries analyses have been proposed for quantifying the predictability of fMRI signals. There are multiple mathematical methods for evaluating complexity in timeseries signals. This study evaluates the optimal parameter settings in seven complexity tests by analyzing mean complexity measures of grey matter (GM) and CSF across multiple complexity tests with different parameter settings and different fMRI acquisition protocols. Furthermore, complexity evaluation was performed on surrogate timeseries data. Overall, Multiscale Sample Entropy, Fuzzy Entropy, and Hurst Exponent consistently showed an increase in mean complexity of GM compared to CSF in original data.Introduction
Over the recent years, the field of resting state fMRI (rs-fMRI) has seen an increase in applications of non-linear timeseries analysis by means of various complexity measures that complement standard linear analyses such as functional connectivity, regional homogeneity or amplitude of low frequency fluctuation1, 2, 3. Our LOFT Complexity Toolbox 2.0 (https://github.com/kayjann/complexity) is designed to estimate complexity of rs-fMRI timeseries data and can perform 7 different complexity tests: Higuchi Fractal Dim (HFD), Hurst Exponent (HurstExp), Lempel-Ziv Complexity (LZC), Approximate Entropy (ApEn), Multiscale Sample Entropy (MSE), Fuzzy Entropy (FuzzyEn), and Permutation Entropy (PE). This study aims at evaluating the optimal parameter selection in these complexity tests as well as the effect of the temporal resolution of rs-fMRI data. A parametric statistical analysis was conducted to compare mean complexity of grey matter (GM) and cerebrospinal fluid (CSF) for different parameter settings of the seven complexity tests. Our assumption is that a good parameter space for a given complexity metric should provide significantly higher complexity in GM than in CSF4.Methods
The study included multiband rs-fMRI scans of 20 subjects (11 females and 9 males) who all underwent rs-fMRI scans with three different sequences (TR = 645 ms; 3 mm isotropic; duration = 10 mins; slices = 40 / TR = 1400 ms; 2 mm isotropic; duration = 10 mins; slices = 64 / TR = 2500 ms; 3 mm isotropic; duration = 5 mins; slices = 38). Subjects were randomly selected from the NKI neuroimaging dataset (Nathan Kline Institute – Rockland Sample). The enhanced NKI project was aimed at creating a large scale (number of participants > 1000) community sample of participants across the lifespan5,6. Pre-processing was performed in CONN-toolbox and included motion realignment, regression of motion parameters and derivatives, regression of CSF and WM signal fluctuations using a-CompCorr, linear drift removal using a high-pass filter, coregistration to individual T1 and normalization to MNI template space. Rs-fMRI scans of one subject were removed from the analysis due to insufficient data quality. The age of the 19-subject sample (original data) was from 15 to 73 (10 females, mean >Signal complexity of rs-fMRI images were measured using the LOFT Complexity Toolbox 2.0. For each parameter in a complexity test, multiple values were chosen (Table 1). Complexity of timeseries signals were measured for each subject scan for all the complexity tests and all the combinations of parameters. Tissue masks for the CSF and GM were based on SPM12 tissue probability maps and the mean complexity value for each tissue type was computed for each subject, scan and complexity measure. The steps were repeated for surrogate data and for all the temporal resolutions. F statistic and differences of mean complexity between groups were calculated using MATLAB repeated measures ANOVA model and post-hoc tests. Statistical significance level was set to p < 0.05.Results
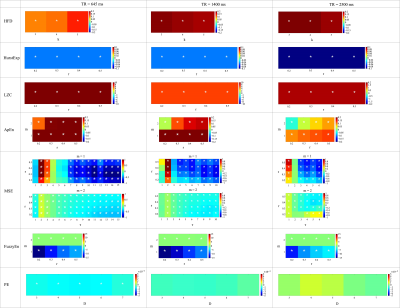
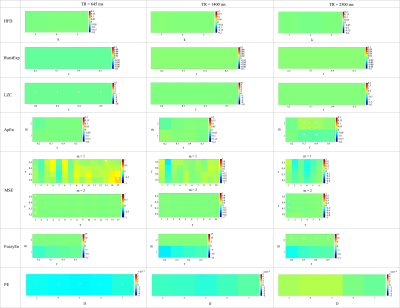
Among 7 complexity tests analyzed in this study, surrogate data of 6 tests (HDF, HurstExp, LZC, ApEn, MSE, and FuzzyEn) showed a significant reduction in differences of means of complexity values between GM and CSF, as compared to original rs-fMRI data (Figure 1 and 2). For permutation entropy, the difference between group means of complexity values among original and surrogate data were indistinguishable. Overall, MSE, FuzzyEn, and HurstExp showed significantly higher mean complexity of GM compared to CSF for all three fMRI protocols. For surrogate data, differences between mean complexity values of GM and CSF were minimal and did not reveal significant differences for most of the parameter settings. For MSE, parameters m = 1 and r = 0.2 - 0.5 provided larger differences in mean complexity between CSF and GM compared to m = 2 parameter setting4. In FuzzyEn, m = 1 parameter setting showed increase in mean complexity values of GM compared to CSF. Overall, MSE demonstrated good discrimination between GM and CSF complexity in a wide parameter space and across all fMRI protocols, while in temporally randomized timeseries there was no significant difference. This was most evident at larger time scales, that represent lower frequency (f) fluctuations (i.e., TR = 645, 1.03 × 10-4 Hz < f < 0.22 Hz; TR = 1400, 7.14 × 10-5 Hz < f < 0.14 Hz; TR = 2500, 6.67 × 10-4 Hz < f < 0.1 Hz).Discussion
From the evaluation of 7 complexity measures in rs-fMRI we recommend using MSE1 at low frequency scales since it showed robust discrimination between GM and CSF signals (assumed to represent mere noise fluctuations) in original timeseries and being sensitive to physiological meaningful data as compared to surrogate timeseries. Notably, MSE is the only method that evaluated complexity at different temporal scales which might provide an inherent advantage. From fMRI connectivity analyses it has been demonstrated that the most meaningful data is in the low frequency range (0.01-0.1Hz) which is consistent with our evaluation7,8. However, further evaluations are necessary specifically due to the small sample size used. In future we will extend this analysis to a voxel-wise analysis with an increased sample size.Acknowledgements
This study was supported by the National Institutes of Health R01AG066711.References
1. Smith RX, Yan L, Wang, DJ. Multiple time scale complexity analysis of resting state FMRI. Brain Imaging Behav, 2014;8(2):284-91.
2. Nezafati M, Temmar H, Keilholz, SD. Functional MRI Signal Complexity Analysis Using Sample Entropy. Front Neurosci, 2020;14:700.
3. Omidvarnia A, Zalesky A, Mansour L S, Van De Ville D, Jackson GD, Pedersen M. Temporal complexity of fMRI is reproducible and correlates with higher order cognition. NeuroImage, 2021;230.
4. Yang AC, Tsai S-J, Lin C-P, Peng C-K. A Strategy to Reduce Bias of Entropy Estimates in Resting-State fMRI Signals. Front. Neurosci. 2018;12:398.
5. Nooner KB, Colcombe SJ, Tobe RH, et al. The NKI-Rockland Sample: A Model for Accelerating the Pace of Discovery Science in Psychiatry. Front Neurosci. 2012; 16;6:152.
6. Tobe RH, MacKay-Brandt A, Lim R, et al. A longitudinal resource for studying connectome development and its psychiatric associations during childhood. Sci Data. 2022; 9, 300.
7. Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34(4):537-41.
8. Lowe MJ, Mock BJ, Sorenson JA. Functional Connectivity in Single and Multislice Echoplanar Imaging Using Resting-State Fluctuations. NeuroImage, 1998;7(2).
Figures