2585
Reproducibility of Pseudocontinuous Arterial Spin Labeling Measured Perfusion in Healthy Volunteers and Glioblastoma Patients1Department of Radiology, University of Texas Southwestern Medical Center, DALLAS, TX, United States, 2Philips Healthcare, Shanghai, China, 3Advanced Imaging Research Center, University of Texas Southwestern Medical Center, DALLAS, TX, United States, 4Department of Neurology, University of Texas Southwestern Medical Center, DALLAS, TX, United States, 5Department of Hematology and Oncology, University of Texas Southwestern Medical Center, DALLAS, TX, United States
Synopsis
Keywords: Arterial spin labelling, Precision & Accuracy, Reproducibility, Precision Accuracy, Perfusion, Cancer, Glioblastoma (GBM), Translational studies
We evaluated the intra-session repeatability and inter-session reproducibility of brain and tumor perfusion measured using 3D pseudo-continuous Arterial spin labeled (PCASL) MRI with TSE based Cartesian acquisition with spiral profile reordering (CASPR) in comparison to 3D pCASL with GRASE in healthy volunteers and glioblastoma (GBM) patients at 3T. 3D pCASL with TSE-CASPR or GRASE provided high intra-session repeatability and 3 weeks inter-session reproducibility in both volunteers and GBM patients. While both readouts generated robust images, TSE-CASPR provided images with reduced distortion particularly in GBM patients and could be a better readout for pCASL measured perfusion in GBM patients.Introduction
Arterial spin labeled (ASL) MRI can provide quantitative perfusion measurements in brain and serves as an early biomarker for treatment evaluation compared to the existing criteria [1, 2]. Due to its non-contrast and non-invasive characteristic, ASL can be potentially used for longitudinal scans to evaluate therapy response assessment in glioblastoma (GBM). Several groups have shown good reproducibility of ASL-MRI measured brain perfusion in healthy volunteers [3-5]. However, for GBM patients, who often have craniotomy, these acquisitions may suffer from B0 inhomogeneities. We recently developed a 3D TSE based Cartesian acquisition with spiral profile reordering (CASPR) combined with pseudo-continuous ASL (pCASL) with increased robustness to B0 inhomogeneities for non-contrast perfusion imaging [6]. The purpose of this study was to evaluate the intra-session repeatability and inter-session reproducibility of brain and tumor perfusion measured using 3D pCASL with TSE-CASPR in comparison to 3D pCASL with GRASE in 20 healthy volunteers and 20 GBM patients at 3T.Methods
Subjects: With IRB approval, 20 healthy volunteers (mean age: 25 ± 2 years) and 20 newly diagnosed GBM patients (mean age: 59 ± 14 years) were recruited. Each healthy volunteer was scanned at two visits (3-week interval) and each visit had two imaging sessions with a 15 mins break in between (Figure 1A). Each session included two runs of 3D pCASL with GRASE and two runs of 3D pCASL with TSE-CASPR (Figure 1B). The 20 GBM patients were imaged before, during, and after chemoradiotherapy for a total of 78 imaging sessions (Figure 1C). At each imaging session, two runs of 3D pCASL with TSE-CASPR readout was performed. In a subset of 33 imaging sessions, two runs of 3D pCASL with GRASE were also acquired. All scans were performed on a 3T MR scanner (Ingenia, Philips Healthcare).Image acquisition: All imaging was performed with a 32-channel head coil. ASL scans were acquired with label duration (LD)/post-label delay (PLD) = 1.8/1.8 s, acquired resolution = 3.5×3.5×6 mm3, reconstructed resolution = 3×3×3 mm3, 4 background suppression pulses and 5 inflow saturation pulses. For TSE-CASPR readout: TR/TE = 6000/14 ms, echo spacing = 2.8 ms, TSE factor = 80, 1 repetition and acquisition time = 3:10 minutes. A M0 image was acquired using the same acquisition parameters in 1:30 minutes. For GRASE: TR/TE = 3955/14 ms, TSE factor = 19, EPI factor = 15, echo spacing = 14.1 ms, 3 repetitions and acquisition time = 4:37 minutes (including the M0). The two readouts had similar acquisition time.
Image analysis: The entire processing pipeline (Figure 2A) for healthy volunteer included format conversion from DICOM to NIfTI followed by skull stripping and co-registration to SRI24 atlas.[7] ASL cerebral blood flow (CBF) maps were calculated based on the ASL consensus paper recommendations.[8] lpba40 labels from SRI24 atlas were used to extract regional perfusion. For GBM patients, all images were co-registered to T1 post contrast images first and tumor ROIs were manually drawn by an experienced radiologist (M. P.). For regional perfusion extraction in GBM patients, all images and tumor ROIs were co-registered to SRI24 atlas such that lpba40 labels could be used to extract normal appearing brain ROIs by subtracting the tumor regions that included whole tumor, cavity, and hemorrhage.
Statistical Analysis: Linear regression and Bland–Altman analyses were performed with GraphPad Prism (GraphPad, San Diego, CA). The reproducibility was measured using intraclass correlation coefficient (ICC). ICC estimates and their 95% confidence intervals (CI) were calculated using SPSS statistical package (SPSS, Chicago, IL) based on a single-measurement, absolute-agreement, 2-way mixed-effects model. Within-subject coefficients of variation (wsCV), defined as the ratio of the standard deviation (SD) of the difference between repeated measurements to the mean of the repeated measurements, were also tabulated.
Results and Discussion
Perfusion images of whole brain were successfully obtained in all healthy volunteers and GBM patients. Representative averaged ASL CBF maps from volunteers are illustrated in Figure 2B for reproducibility analysis among three conditions. For GBM patients, however, only intra-session repeatability was evaluated since brain perfusion changes at different time points could be altered by chemoradiation, tumor progression, and/or regression.Corresponding linear regression and Bland-Altman analyses of mean CBF values among normal appearing ROIs and tumor regions are shown in Figures 3 and 4 for volunteers and patients respectively. Both suggested excellent correlation (R2 > 0.9) and minimal bias (bias < 1) for intra-session repeatability, which has been further validated by ICC values (>0.90) and wsCVs (<10%) (Figure 5A). On the other hand, lower reproducibility was observed among all analyses for 15 mins and 3 weeks inter-session in volunteers as expected. The 15 mins inter-session reproducibility of TSE-CASPR was much poorer compared to that of GRASE, which would require further investigation for underlying mechanisms. However, the intra-session repeatability and 3 weeks inter-session reproducibility, which are more relevant for clinical analyses showed good correlation for both TSE-CASPR and GRASE.
Conclusion
3D pCASL with TSE-CASPR or GRASE provided high intra-session repeatability and 3 weeks inter-session reproducibility in both healthy volunteers and GBM patients at 3 T. While both readouts generated robust images, TSE-CASPR provided images with reduced distortion particularly in GBM patients (Figure 5B) and could be a better readout for pCASL measured perfusion in GBM patients [9].Acknowledgements
This work was supported by NIH/NCI grant U01CA207091. The authors thank Kelli Key, PhD, Abey Thomas, RT(MR), Courtney Dawson, RT(MR), Michael Fulkerson, AS, LVN, and Sydney Haldeman, MPH, for their help in human imaging, and Ben Wagner, MSEE, for his help with image database and analysis routines. The authors would also like to thank all patients and healthy volunteers for their participation in this study.
References
1. Eisenhauer, E.A., et al., New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer, 2009. 45(2): p. 228-47.
2. Wen, P.Y., et al., Response Assessment in Neuro-Oncology Clinical Trials. J Clin Oncol, 2017. 35(21): p. 2439-2449.
3. Wu, B., et al., Intra- and interscanner reliability and reproducibility of 3D whole-brain pseudo-continuous arterial spin-labeling MR perfusion at 3T. J Magn Reson Imaging, 2014. 39(2): p. 402-9.
4. Mutsaerts, H.J., et al., Inter-vendor reproducibility of pseudo-continuous arterial spin labeling at 3 Tesla. PLoS One, 2014. 9(8): p. e104108.
5. Wang, Y., et al., Regional reproducibility of pulsed arterial spin labeling perfusion imaging at 3T. Neuroimage, 2011. 54(2): p. 1188-95.
6. Greer, J.S., et al., Robust pCASL perfusion imaging using a 3D Cartesian acquisition with spiral profile reordering (CASPR). Magn Reson Med, 2019. 82(5): p. 1713-1724.
7. Rohlfing, T., et al., The SRI24 multichannel atlas of normal adult human brain structure. Hum Brain Mapp, 2010. 31(5): p. 798-819.
8. Alsop, D.C., et al., Recommended Implementation of Arterial Spin-Labeled Perfusion MRI for Clinical Applications: A Consensus of the ISMRM Perfusion Study Group and the European Consortium for ASL in Dementia. Magnetic Resonance in Medicine, 2015. 73(1): p. 102-116.
9. Zhou, L., et al., Intrasession Reliability of Arterial Spin-Labeled MRI-Measured Noncontrast Perfusion in Glioblastoma at 3 T. Tomography, 2020. 6(2): p. 139-147.
Figures
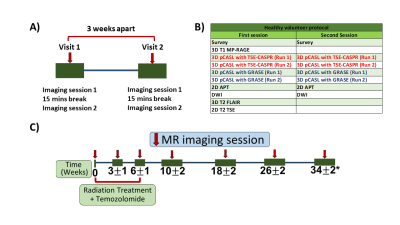
Figure 1: MR scan timelines and imaging protocol for healthy volunteers (A, B) and GBM patients (C). A) Each healthy volunteer was recruited for two visits at three weeks interval. Each visit included two imaging sessions with a 15 mins break. B) The MR imaging protocol for each visit of healthy volunteers. C) The MR scan timeline for GBM patients, with MR scans acquired before, during, and after the radiation treatment.
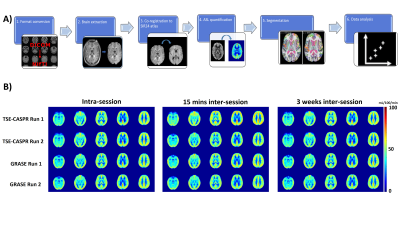
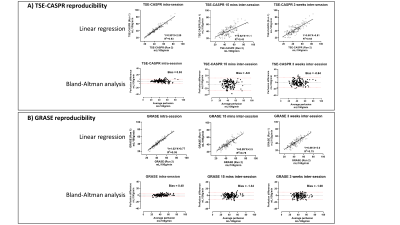
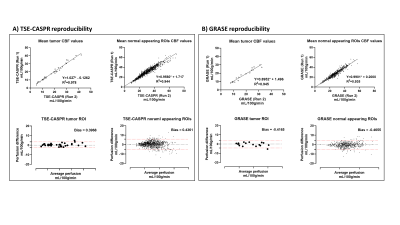
Figure 4: Linear regression and Bland-Altman analyses of 2 runs of CBF measurements in mL/100g/min using 3D pCASL with TSE-CASPR (Figure 4A) and GRASE (Figure 4B) for tumor areas (left) and normal appearing ROIs (right) among GBM patients for intra-session reproducibility.
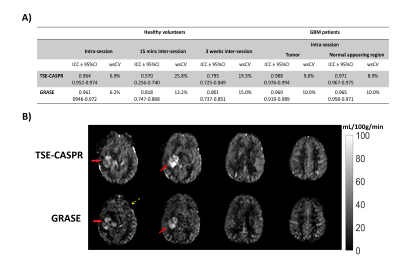
Figure 5: A) ICC and wsCV for CBF measurements between 2 Runs of 3D pCASL with TSE-CASPR and GRASE for healthy volunteers (intra-session, 15 mins and 3 weeks inter-session reproducibility) and GBM patients (intra-session reproducibility only). B) Representative CBF maps of a GBM patient acquired for 3D pCASL with TSE-CASPR (top row) and with GRASE (bottom row). CBF maps are visually very similar, including tumor (solid arrow). 3D TSE-CASPR images provided better tumor to background gray matter contrast and are more robust to B0 inhomogeneities compared with 3D GRASE (dashed arrow).