2576
The ASL Challenge Reproduction Study: Replicating Findings from The OSIPI-ISMRM ASL MRI Challenge
Channelle Tham1, Ahmed Abdalle2, Joseph G Woods3, Maria-Eleni Dounavi4, Andre Pascoal5, and Udunna Anazodo6
1Department of Cognitive Neuroscience, Radboud University, Nijmegen, Netherlands, 2Schulich School of Medicine and Dentistry, Western University, London, ON, Canada, 3Wellcome Centre for Integrated Neuroimaging, FMRIB, Nuffield Department of Clinical Neuroscience, University of Oxford, Oxford, United Kingdom, 4Department of Psychiatry, University of Cambridge, Cambridge, United Kingdom, 5Instituto de Radiologia e Departamento de Radiologia e Oncologia, University of Sao Paulo, Sao Paulo, Brazil, 6Montreal Neurological Institute, McGill University, Montreal, QC, Canada
1Department of Cognitive Neuroscience, Radboud University, Nijmegen, Netherlands, 2Schulich School of Medicine and Dentistry, Western University, London, ON, Canada, 3Wellcome Centre for Integrated Neuroimaging, FMRIB, Nuffield Department of Clinical Neuroscience, University of Oxford, Oxford, United Kingdom, 4Department of Psychiatry, University of Cambridge, Cambridge, United Kingdom, 5Instituto de Radiologia e Departamento de Radiologia e Oncologia, University of Sao Paulo, Sao Paulo, Brazil, 6Montreal Neurological Institute, McGill University, Montreal, QC, Canada
Synopsis
Keywords: Arterial spin labelling, Challenges, Reproducibility
While the recommendation for ASL acquisition is widely adopted to standardize quantification of cerebral blood flow (CBF), ASL analysis still produces wide variability in CBF estimates. The recent OSIPI-ISMRM ASL MRI Challenge highlighted how differences in image processing contribute to CBF variability. Here, we explored the replicability of ASL image processing approaches when used as documented to quantify CBF. The results of 5 of the 8 ASL Challenge teams were replicated and compared to their submitted results. The overall errors in global mean CBF were 0.03-5 times larger in the replicated results, demonstrating the need for ASL analysis standards.INTRODUCTION
Arterial Spin Labeling (ASL) is a widely used MRI technique for quantitative evaluation of physiological measures linked to blood perfusion in the brain. While several ASL acquisition and readout schemes exist and continue to be areas of active research, the pseudo-continuous (PCASL) labeling scheme is the recommended 1 approach for clinical applications. However, clinical adoption of PCASL is limited by standardization of acquisition parameters and analysis methods. Building on the recent consensus guideline for PCASL acquisition 1, the Open Science Initiative for Perfusion Imaging (OSIPI) and the ISMRM ASL MRI Challenge 2,3 was organized to provide a community-driven consensus on best practice for PCASL image processing and analysis, as well as understand sources of variability in cerebral blood flow (CBF) measures. In this work, we utilized the 2022-2023 ISMRM Repeat It with Me: Reproducibility Team Challenge 4 to further explore the repeatability of image processing and analysis pipelines for PCASL CBF measurement. We replicated the CBF results submitted for the OSIPI-ISMRM ASL MRI Challenge (henceforth ASL Challenge) and compared measurement errors to ground truth CBF.METHODS
Eight teams submitted CBF maps and documentation of their image processing and analysis steps. The Challenge dataset 2,3 consisting of nine synthetic data simulated using an ASL digital reference object (DRO) to measure CBF in normal and diseased states was used for this reproduction study. The data contained four-dimensional (4D) time-series of PCASL control-label pairs, a calibration (M0) image, and a high-resolution T1-weighted anatomical image. All data used in this study were obtained from the Open Science Framework (OSF) repository. Submitted CBF maps and documentation were provided by the ASL Challenge scoring team. Details of the ASL data parameters, simulated conditions and challenge evaluation are on the challenge website 2. To replicate results, each participating team’s pipeline and analysis steps were used as documented to analyze the nine datasets. Teams were contacted for clarification when errors in deploying the pipeline/analysis scripts were encountered. The submitted CBF were accessed after generating a replication of CBF maps. To evaluate the replicability of the analysis pipelines, the error in mean absolute GM CBF across all voxels in the brain were calculated as the difference in the replicated or submitted CBF estimates and the ground truth using MATLAB scripts developed by the ASL Challenge scoring team. For both the ASL Challenge and the Repeat with Me Challenge DRO simulation parameters were withheld during data analysis to reduce bias. The replication was performed by two trainees (CT and AA) who have beginner level skills (~5 months) in ASL analysis.RESULTS
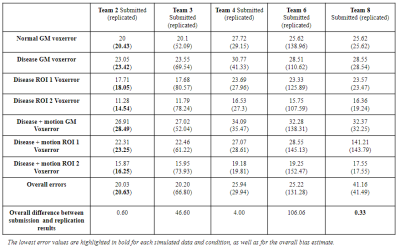
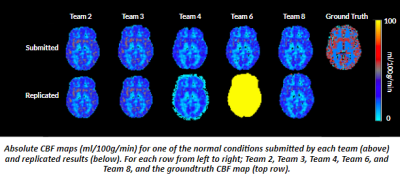
Five of the eight submitted pipelines were replicated to generate CBF maps. Three submissions were not replicated because of unclear documentation, errors in executing analysis scripts or the pipeline was too crude and could not be replicated without extensive assistance from the developer. The pipelines replicated are shown in Table 1. Overall, the absolute mean global CBF errors in submitted and replicated results ranged from 20.3 to 131.28 ml/100g/min, across the 5 team submissions (Table 2). Axial slices of CBF maps from a normal synthetic dataset are shown in Figure 1 for each submission and corresponding replicated result.DISCUSSION
This study was primarily designed to evaluate the replicability of ASL analysis methods as an approach to explore the reliability of reproducing ASL analysis of CBF measures conducted using the same conditions and experimental setup (same ground truth data). Overall, the absolute error in global CBF across voxels in gray matter in normal and diseased conditions were quite large in the replicated results and more importantly, larger than the errors observed from submissions to the ASL Challenge. As noted in the ASL Challenge findings, the wide variability in CBF values may be attributed to differences in image processing strategies. With the exception of Team 2 , 4, and 8, the absolute global CBF gray matter CBF reproduced were 3-5 times higher than the original values from submitted CBF maps. Although the teams were consulted during the analysis to clarify analysis steps or debug codes in an attempt to replicate results as closely as submitted, the larger error margins in the replicated results demonstrate ongoing challenges with reproducing CBF measures using current ASL analysis practices. Based on the lowest difference in CBF bias in replicated results, a case could be made for the higher replicability of widely used, open-sourced, and long standing analysis methods (Team 2, 4, and 8) which tend to have better documentation and less execution errors/bugs. Although this study evaluated a relative small proportion of ASL analysis methods, the findings of wide variation in measured CBF and lower replicability in 50% of evaluated submissions, demonstrate not only the significance of reproducible science, but underscored further the need to standardize ASL analysis pipelines to ensure reproducible CBF measures for research and clinical environments.CONCLUSION
In general, the analysis of PCASL data to quantify cerebral blood flow remains an important area of interest in advancing ASL towards clinical adoption. This work complements the OSIPI-ISMRM ASL MRI Challenge by further demonstrating the need for recommended standards for image processing and analysis steps.Acknowledgements
The authors thank Steve Sourbron, Laura Bell and Henk-Jan Mutsaerts, past and current chairs of the Open Science Initiative for Perfusion Imaging (OSIPI) for establishing an open science imaging ecosystem for evaluating replicability of MRI methods. The authors are grateful to the OSIPI Task Force 6.1: ASL Challenges members for organizing the OSIPI-ISMRM ASL MRI Challenge and the Reproducibility Team Challenge organizers (Drs Laura Bortolotti, Sophie Schauman, and Maria Eugenia Caligiuri) for the opportunity to perform this study.References
1. Alsop DC, Detre JA, Golay X, et al. Recommended implementation of arterial spin-labeled perfusion MRI for clinical applications: A consensus of the ISMRM perfusion study group and the European consortium for ASL in dementia. Magn Reson Med. 2015;73(1):102-116.2. The Open Science Initiative for Perfusion Imaging (OSIPI) ASL Challenge. https://challenge.ismrm.org/forums/topic/osipi-asl-challenge/
3. Anazodo U, Pinto J, McConnell FK, et al. The Open Science Initiative for Perfusion Imaging (OSIPI): Results from the ASL MRI Challenge. Proceedings of the Joint ISMRM-ESMRMB Annual Meeting (2022).
4. Repeat It With Me: Reproducibility Team Challenge. https://challenge.ismrm.org
5. Oxford_asl analysis tool. https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/oxford_asl
6. Quantiphyse. https://quantiphyse.readthedocs.io/en/latest/asl/asl_analysis.html
7. Chappell MA, Groves AR, Whitcher B, Woolrich MW. Variational Bayesian inference for a nonlinear forward model. IEEE Trans. Signal Process. 2009;57:223–236
8. Mutsaerts HJMM, Petr J, Groot P, et al. ExploreASL: an image processing pipeline for multi-center ASL perfusion MRI studies. Neuroimage 2020;219:117031
9. ASL MRI Cloud. https://braingps.mricloud.org/docs/Manual_ASL_processing.v2.pdf
10. Bron EE, Steketee RME, Houston GC, et al. Diagnostic classification of arterial spin labeling and structural MRI in presenile early stage dementia. Hum. Brain Mapp. 2014;35:4916–4931
11. LOFT ASL toolbox https://github.com/chenyang9526/LOFT_ASL_toolbox
DOI: https://doi.org/10.58530/2023/2576