2563
A Physiologically-Decomposed DWI machine-learning model improves prediction of response to NAC treatment in invasive breast cancer
Maya Gilad1 and Moti Freiman2
1Efi Arazi School of Computer Science, Reichman University, Herzliya, Israel, 2Faculty of Biomedical Engineering, Technion-Israel Institute of Technology, Haifa, Israel
1Efi Arazi School of Computer Science, Reichman University, Herzliya, Israel, 2Faculty of Biomedical Engineering, Technion-Israel Institute of Technology, Haifa, Israel
Synopsis
Keywords: Breast, Diffusion/other diffusion imaging techniques
Early prediction of pathological complete response (pCR) following neoadjuvant chemotherapy for breast cancer plays a critical role in surgical planning and optimizing treatment strategies. Recently, machine and deep-learning based methods were suggested for early pCR prediction from multi-parametric MRI data with moderate success. We introduce PD-DWI, a physiologically-decomposed DWI machine-learning model to predict pCR from DWI and clinical data. Our model first decomposes the raw DWI data into the various physiological cues that are influencing the DWI signal and then uses the decomposed data, in addition to clinical variables, as the input features of a radiomics-based XGBoost model.Introduction
Neoadjuvant chemotherapy (NAC) is a pre-operative standard-of-care for invasive breast cancer and has been shown to improve breast cancer treatment effectiveness and has been associated with better survival rates1,6. Pathological complete response (pCR), the absence of residual invasive disease in either breast or axillary lymph nodes, is used to assess patient response to NAC. While notable pCR rates have been reported even in the most aggressive tumors, approximately 20% of breast cancers are resistant to NAC; hence, early response prediction of pCR might enable efficient selection and, when needed, alteration of treatment strategies and improve surgical planning2,7.Our goal is to investigate the value of a Physiologically-Decomposed DWI machine-learning model in predicting pCR to NAC treatment in invasive breast cancer patients.
Methods
Patient cohortThe Breast Multiparametric MRI (mpMRI) for the prediction of NAC Response (BMMR2) Challenge cohort was used3-5. Data included standardized longitudinal mpMRI datasets as well as clinical characteristics of 191 breast cancer patients curated from the prospective multicenter ACRIN6698 study by challenge organizers. mpMRI data (DWI, DCE-MRI, T2W) was acquired at pre-treatment, early treatment, and mid-treatment. Diffusion gradients were applied in three orthogonal directions by using diffusion weightings (b-values) of 0, 100, 600, and 800 s/mm2.
Patients were split into training (60%) and test (40%) groups and reference histopathological pCR classification was available only for the training group.
Physiologically-Decomposed DWI
We calculated ADC maps from different ranges of b-values using the mono-exponential decay model: 1) ADC0-100, 2) ADC0-800, and 3) ADC100-800. A pseudo-diffusion fraction (F) map was calculated from ADC0-100 and ADC0-800 (See Fig. 1-2).
Machine learning model
We developed a physiologically-decomposed XGBoost classification model combining clinical variables and 3D radiomic features extracted from the ADC and F maps (see Fig. 3).
To mitigate the large number of features compared to the size of the training group, we’ve included multiple feature selection techniques. We selected a subset of features based on the F-ANOVA score in addition to adjusting the max-depth and min-child-weight parameters of XGBoost; hence, the number of used features is limited and each selected feature is representative of the group. We’ve also used the subsample parameter to increase the model’s robustness.
Evaluation
Our model performance was assessed objectively on the testing group by challenge organizers using the Area Under Curve (AUC) metric as the evaluation metric.
We created a baseline model based on ADC0-800 and SER maps to asses DWI as a single imaging biomarker for pCR prediction. The significance of physiologically decomposing the DWI signal was assessed by comparing the PD-DWI model to models with different ranges of ADC maps and using only F map. In addition, we conducted a longitudinal analysis of the PD-DWI model compared to the baseline model to assess its performance for early prediction of pCR following NAC.
Results
BMMR2 challenge performance comparisonPD-DWI presents superior pCR prediction over the best results published as part of BMMR2 official challenge (see Fig. 4). To the best of our knowledge, our model has the best pCR prediction performance for the BMMR2 dataset.
Relation between DWI signal attenuation decay and pCR prediction
PD-DWI model achieved a higher AUC score than the ADC-only models and the F-only model. Amongst the ADC-only models, ADC0-800 had the highest AUC score, followed by ADC0-100. AUC scores of ADC100-800 and F-only model scores were relatively similar.
Upon the release of test group labels, we evaluated the statistical significance of PD-DWI model compared to ADC-only models and the F-only model. PD-DWI model achieved the best Cohen’s Kappa (κ) score whilst ADC0-800 was second to last. Furthermore, PD-DWI's improved performance over the F-only model is statistically significant (Sensitivity test, 0.65, p < 0.05) and also compared to the baseline model (Sensitivity test, 0.6, p < 0.05).
Early prediction of pCR over the course of NAC
As expected, both models’ pCR predictions improve given additional imaging data acquired during the course of the treatment (see Fig. 5). PD-DWI model outperforms the baseline model on both pre-NAC and min-NAC. In addition, PD-DWI model performance using only pre-NAC data achieved a higher pCR prediction performance compared to the BMMR2 benchmark which used all time points as provided by challenge organizers (0.7928 vs 0.78).
Discussion
PD-DWI's improved performance over ADC-only and F-only models suggests that there are changes in both cellular density and blood flow in the micro-capillary network as a response to NAC.Findings from our longitudinal analysis suggest that changes in pseudo-diffusion and pseudo-diffusion fraction are more evident compared to changes in overall ADC values and SER values during the course of the treatment. Whilst both models performed about the same at early-NAC, PD-DWI model has the advantage of not relying on DCE-MRI.
Conclusion
PD-DWI model can effectively enable treatment optimization by predicting pCR from mpMRI data before and during NAC treatment. Further, our results suggest that pCR prediction can be achieved using DWI data solely. Therefore, has the potential to eliminate the need for DCE-MRI scans which require contrast agent injection and longer acquisition time. Moreover, decomposing DWI data even from a small number of b-values to physiological components can provide meaningful information to improve ML models' performance.Acknowledgements
We thank the BMMR2 challenge organizers for the data sharing and post-challenge useful discussions.References
- A. Bhushan, A. Gonsalves, and J. U. Menon. Current state of breast cancer diagnosis,treatment, and theranostics. Pharmaceutics, 13, 2021
- D. Song, X. Man, M. Jin, Q. Li, H. Wang, and Y. Du. A decision-making supporting prediction method for breast cancer neoadjuvant chemotherapy. Frontiers in Oncology, 10,2021.
- K. Clark, B. Vendt, K. Smith, J. Freymann, J. Kirby, P. Koppel, S. Moore, S. Phillips,D. Maffitt, M. Pringle, L. Tarbox, and F. Prior. The cancer imaging archive (tcia): Main-taining and operating a public information repository, Jul 2013.
- D. C. Newitt, S. C. Partridge, Z. Zhang, J. Gibbs, T. Chenevert, M. Rosen, P. Bolan,H. Marques, J. Romanoff, L. Cimino, B. N. Joe, H. Umphrey, H. Ojeda-Fournier, B. Dogan,K. Y. Oh, H. Abe, J. Drukteinis, L. J. Esserman, and N. M. Hylton. Acrin 6698/i-spy2breast dwi, 2021.
- S. C. Partridge, Z. Zhang, D. C. Newitt, J. E. Gibbs, T. L. Chenevert, M. A. Rosen, P. J.Bolan, H. S. Marques, J. Romanoff, L. Cimino, B. N. Joe, H. R. Umphrey, H. Ojeda-Fournier, B. Dogan, K. Oh, H. Abe, J. S. Drukteinis, L. J. Esserman, and N. M. Hylton.Diffusion-weighted mri findings predict pathologic response in neoadjuvant treatment ofbreast cancer: The acrin 6698 multicenter trial. Radiology, 289, 2018.
- M. Banaie, H. Soltanian-Zadeh, H. R. Saligheh-Rad, and M. Gity. Spatiotemporal features of dce-mri for breast cancer diagnosis. Computer Methods and Programs in Biomedicine,155, 2018.
- E. H. Cain, A. Saha, M. R. Harowicz, J. R. Marks, P. K. Marcom, and M. A. Mazurowski.Multivariate machine learning models for prediction of pathologic response to neoadjuvant therapy in breast cancer using mri features: a study using an independent validation set.Breast Cancer Research and Treatment, 173, 2019.
Figures
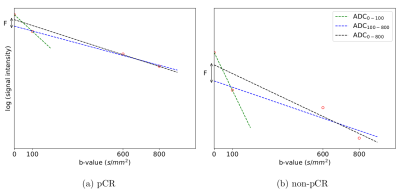
Figure 1: DWI signal decay as a function of the b-value along with the different ADC models of representative pCR and Non pCR patients.
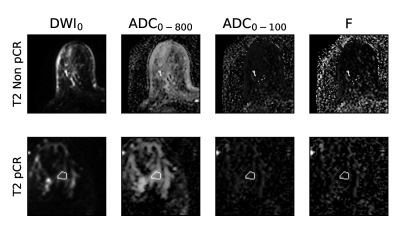
Figure 2: DWI data of pCR and non-PCR patients at T2 (mid-NAC). The white contour represents the tumor segmentation used for analysis as given by challenge organizers.
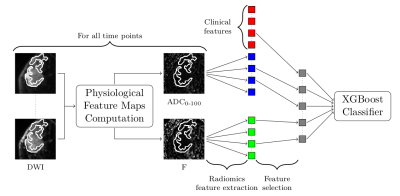
Figure 3: PD-DWI Model architecture: The acquired DWI data at each time-point is decomposed into the different physiological cues. 3D Radiomics features are extracted from the segmented tumor in ADC and F maps and categorical clinical data are concatenated. Then a feature selection process is applied to select the most informative features. Finally, the selected features are fed into an XGBoost classifier.
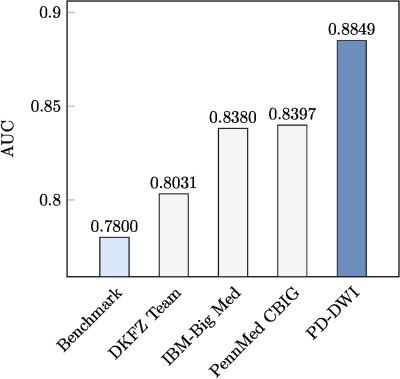
Figure 4: BMMR2 Challenge performance comparison at time-point T2 (mid-NAC).
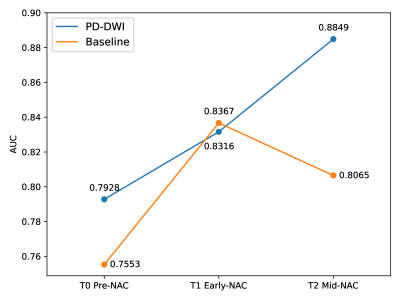
Figure 5: Model performance at the different phases of NAC treatment
DOI: https://doi.org/10.58530/2023/2563