2544
3D Multi-echo spiral acquisition with model-based reconstruction for fMRI
Zidan Yu1, Christoph Rettenmeier1, and V. Andrew Stenger1
1Department of Medicine, University of Hawaii, Honolulu, HI, United States
1Department of Medicine, University of Hawaii, Honolulu, HI, United States
Synopsis
Keywords: Data Acquisition, fMRI
Multi-echo fMRI is of current interest due to its potential for more accurate BOLD contrast with reduced artifacts. Furthermore, spiral trajectories are advantageous for fMRI because of high sampling efficiency. However, images are susceptible to blurring due to B0 inhomogeneity. In this study, we present a 3D multi-echo spiral sequence with a model-based reconstruction method that combines under sampled spiral data from different echoes for increased sampling efficiency and reduced B0 artifacts. Multi-echo and single-echo spiral fMRI data including T2* maps were acquired and compared at 3T demonstrating the method.Introduction
Multi-echo fMRI is of current interest due to its potential for more accurate BOLD contrast with reduced artifacts1. Furthermore, spiral trajectories are advantageous for fMRI because of high sampling efficiency. However, images are susceptible to blurring due to B0 inhomogeneity2. In this study, we present a 3D multi-echo spiral sequence with a model-based reconstruction method that combines under sampled spiral data from different echoes for increased sampling efficiency and reduced B0 artifacts. Multi-echo and single-echo spiral fMRI data including T2* maps were acquired and compared at 3T demonstrating the method.Methods
A prototype multi-echo 3D GRE sequence was designed with four spiral readouts after each RF excitation (22°) in every TR (75ms). As shown in Figure 1, each spiral readout was under sampled with R = 2 and a 180° rotation was applied between each. We used an iterative reconstruction method to model the images at different TEs based on the signal equation that employs a B0 field map3, R2* map, and coil sensitivities4 acquired from a separate calibration scan. Every sequential pair of under sampled spiral k-space data were combined with the model-based reconstruction using the TE value of the first data set in the pair. As a result, we can get multi-echo datasets for three echoes from the four under sampled spiral acquisitions in the same time as a standard single-echo spiral fMRI acquisition. A NUFFT5 was applied to the spiral k-space samples and a limited-memory BFGS (Poblano toolbox) algorithm was used for the optimization of the model. A matched filter reconstruction was applied to the same dataset by simply combining every two spirals for comparison. A fully sampled SENSE map was used in this case.In-vivo fMRI experiments were also performed using the multi-echo sequence and a standard single-echo spiral matched for 3mm isotropic resolution and 19.2cm FOV with a 25ms TE for comparison. The protocol for the multi-echo sequence was set with 24 slices and TEs were set at 2.4ms, 14.65ms, 26.90ms, and 39.15ms for each echo. Thus, the model-based reconstruction results have echoes at TE = 2.4ms, 14.65ms, 26.90ms. The fMRI tasks consisted of 6 blocks of 20s flickering checkerboard for visual stimulation, separated by 20s resting periods leading to a total duration of the paradigm of 4:20 min. BOLD analyses were carried out using a general linear model with a canonical hemodynamic response function after removal of first and second order temporal trends in the data. No spatial smoothing, masking, or corrections was applied. All scans were acquired on a 3T Siemens Prisma scanner and reconstructions were done offline using Matlab.
Results
As shown in Figure 2, the images from the standard matched filter reconstruction have obvious phase discrepancies. The model-based reconstruction however performs much better when generating the multi-echo results. The multi-echo method also mitigates the B0 distortion around the sinuses in the lower slices when comparing to the single-echo spiral results in Figure 3. In addition, the edge of the brain can be clearly depicted by the multi-echo sequence where the b>Discussion & Conclusion
Our multi-echo 3D spiral sequence in combination with the model-based reconstruction can acquire multiple echoes of comparable resolution as a single-echo scan of the same duration for T2* weighted fMRI. Furthermore, the multi-echo spiral has significantly reduced B0 artifacts. This is due to both the shorter readouts of the multi-echo scan and the use of B0 information in the reconstruction. The fMRI results did show reduced BOLD activation. However no multi-echo optimization such as averaging or contrast weighting was applied and needs further investigation.Acknowledgements
This project was supported by the NIH grants 1P20GM139753-01A1) and R01 EB023618.References
1. Kundu, P. et al. Multi-echo fMRI: A review of applications in fMRI denoising and analysis of BOLD signals. NeuroImage 2017;154:59–80.
2. Glover, G. H. Spiral imaging in fMRI. NeuroImage 2012;62:706–712.
3. Dymerska B, Eckstein K, Bachrata B, et al. Phase unwrapping with a rapid opensource minimum spanning tree algorithm (ROMEO)[J]. Magnetic resonance in medicine, 2021;85(4):2294-2308.
4. Walsh DO, Gmitro AF, Marcellin MW. Adaptive reconstruction of phased array MR imagery. Magnetic resonance in medicine, 2000;43(5):682-690.
5. Fessler JA, Sutton BP. Nonuniform fast Fourier transforms using min-max interpolation. IEEE Transactions on Signal Processing 2003;51(2):560-574
Figures
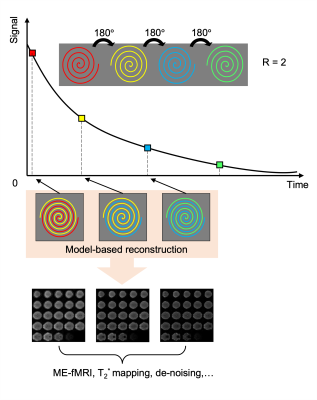
Figure 1: The multi-echo spiral sequence design and the illustration of the reconstruction method.
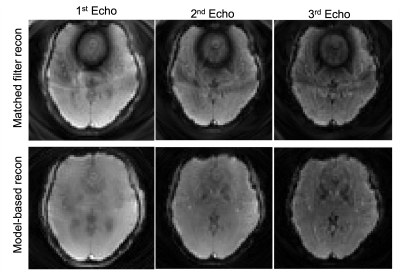
Figure 2: Reconstruction results: the 1st row shows the multi-echo images from slice #18 using matched filter reconstruction. We just simply combined every 2 spirals. The 2nd row shows the model-based reconstruction results.
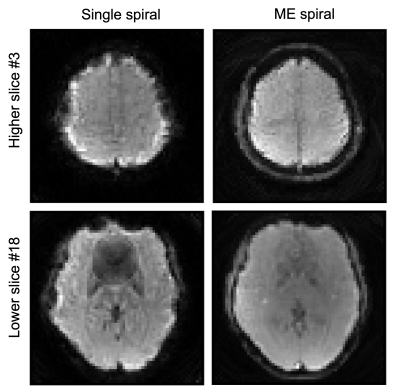
Figure 3: Comparison of the structural images between single spiral and multi-echo spiral datasets. Severe B0 artifacts can be observed in the single spiral images.
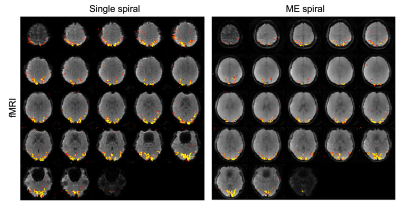
Figure 4: fMRI results from single-echo spiral scan and the multi-echo scan.
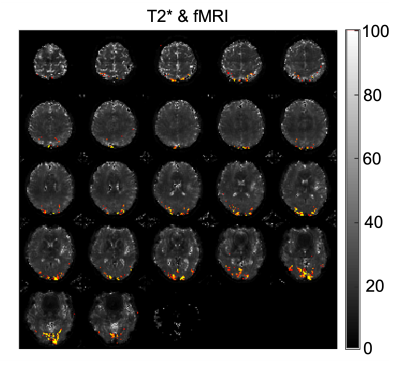
Figure 5: The T2* results we generated from the multi-echo fMRI scan and the BOLD signal we got from the whole time series of T2* maps.
DOI: https://doi.org/10.58530/2023/2544