2542
Noise Reduction for Task-Based Functional Magnetic Resonance Imaging using NORDIC and KWIA1Mark and Mary Stevens Neuroimaging and Informatics Institute, University of Southern California, Los Angeles, CA, United States, 2Monash Biomedical Imaging, Department of Data Science & AI, Faculty of IT, Monash University, Clayton, Australia
Synopsis
Keywords: Image Reconstruction, Image Reconstruction
We quantitatively evaluate the performance of NORDIC and KWIA methods in denoising of high-resolution task-fMRI data from 7T in comparison to the standard manufacturer reconstruction. We found that both techniques improve single run statistics as well as improved consistency across runs. Analysis of tSNR confirmed improved data quality in the time domain that facilitates time-series analysis for task-fMRI.Introduction
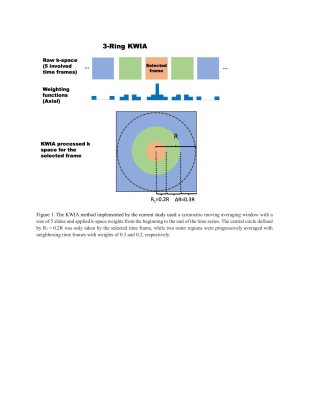
Functional magnetic resonance imaging (fMRI) has found great utility for a wide range of neuroscientific and clinical applications1-3. However, high-resolution fMRI, which is required for improved delineation of fine brain structures and connectomics, is hampered by its low signal-to-noise ratio (SNR). Recently, Noise Reduction with Distribution Corrected (NORDIC) denoising method was proposed to suppress the noise components of image series that cannot be distinguished from thermal noise4. Our group also developed a denoising method termed k-space weighted average image (KWIA)5. KWIA performs spatiotemporal filtering of image time series by taking advantage of the characteristics of k-space data. In this work, we quantitatively evaluate the performance of NORDIC and KWIA methods in denoising by comparing them to the standard manufacturer reconstruction for task-based high-resolution fMRI datasets.Method
Imaging Experiments: Two volunteers participated in repeated visual task fMRI experiments on a 7T Siemens Magnetom Terra using 32 channel head coil (Nova Medical, Wilmington, MA). A visual checkerboard block design was displayed on an MR compatible screen. The task included four blocks, each starting with displaying on the black background a flickering checkerboard (8Hz) for 20s followed by a white fixation cross for another 20s. We ran this task for four times, each with the duration of 160s. For Subject 1, a total of 90 functional images were collected for each of the four runs with a HCP style fMRI sequence with 1mm isotropic resolution (192X192 matrix, TR=2,000ms; TE=21ms; flip angle=73°; number of slices=125, MB5 GRAPPA2). The structural T1-weighted image was acquired using the MPRAGE sequence (320X300 matrix, TR=4,300ms; TE=2.27ms; flip angle=4°; voxel size=0.75X0.75X0.75mm3, number of slices=192). For Subjects 2, all the scanning parameters were the same with Subject 1 except functional images were collected with 0.8mm isotropic resolution (240X240 matrix, TE=24.2 ms; number of slices=116). All image reconstructions and denoising were implemented in MATLAB (R2020b; the Mathworks, Inc., Natick, MA, USA), for off-line reconstruction and denoising using a Linux (CentOS) workstation with 48-Core Intel(R) Xeon(R) Gold 6248R CPU @ 3.00GHz and 512-GB of Memory. Each fMRI run was separately reconstructed with GRAPPA parallel imaging reconstruction algorithm (STANDARD)6, then KWIA5 denoising (Figure 1) and NORDIC7 were applied to the previous GRAPPA reconstructed images, respectively.Data Analysis: Functional MRI data were preprocessed and analyzed using Statistical Parametric Mapping (SPM) across the four fMRI runs for each reconstruction method. Reconstructed fMRI timeseries were realigned to correct motion and high-pass filtered and then coregistered to the anatomical image. No spatial smoothing was performed. Preprocessed images were then analyzed separately for each run and reconstruction technique using a General Linear Model with predictor for visual stimulation and nuisance regressors for motion parameters. Second level analyses calculating a one-sample t-statistic across the 4 runs and also for each run in each reconstruction modality revealed consistency end extent of significant voxels. Based on the t-statistic map from the Standard reconstruction two regions of interests for left and right visual cortex were generated and average fMRI timeseries data from those ROIs were extracted from each run and reconstruction to compute the respective temporal SNR (tSNR). A one-way analysis of variance (ANOVA) was conducted to evaluate tSNR differences for the three reconstruction methods.
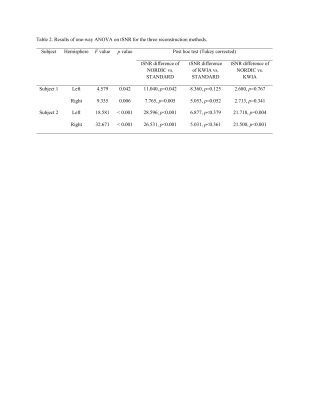
Results
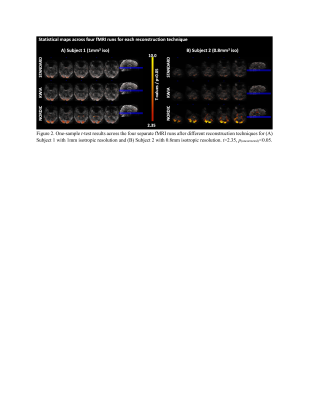
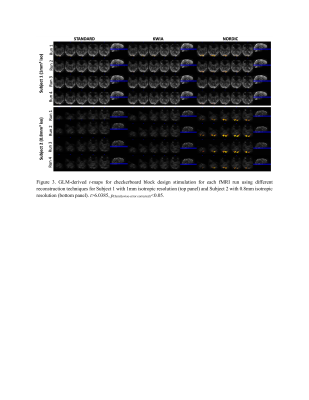
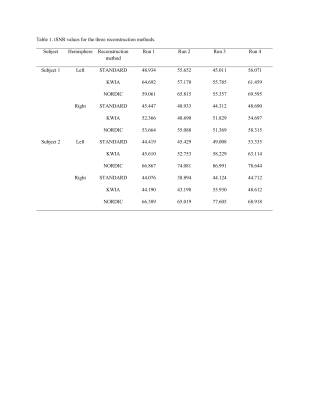
NORDIC outperformed KWIA and STANDARD as reflected by more consistent and larger extend of visual cortex activation in t-maps across the four fMRI runs (t>2.35, p(uncorrected)<0.05, Figure 2) and for each fMRI run (t>6.0385, p(familywise-error corrected)<0.05, Figure 3). Numbers of voxels in each group test for Subject 1, STANDARD 21795, KWIA 15263 and NORDIC 81698 and for Subject 2, STANDARD 3493, KWIA 6040 and NORDIC 40341* (*value for NORDIC is at p<0.01). The ANOVA analysis for tSNR revealed a significant effect of reconstruction method in the V1 region (For Subject 1, left side: F=4.579, p=0.042; right side: F=9.335, p=0.006. For Subject 2, left side: F=18.581, p<0.001; right side: F=32.671, p<0.001; Table 1&2). The post hoc analysis with Tukey correction indicated for Subject 1, NORDIC had significantly higher tSNR than STANDARD (Left side: difference of tSNR=11.040, p=0.042; right side: difference of tSNR=7.765, p=0.005); KWIA had a marginally higher tSNR than STANDARD on the right side (difference of tSNR=5.053, p=0.052). For Subject 2, NORDIC had significantly higher tSNR than STANDARD (Left side: difference of tSNR=28.596, p<0.001; right side: difference of tSNR=32.671, p<0.001) and KWIA (Left side: difference of tSNR=21.718, p=0.004; right side: difference of tSNR=21.500, p<0.001).Conclusion
We compared two different noise reduction reconstruction techniques against the manufacturer standard reconstruction. We found that both techniques improve single run statistics as well as improved consistency across runs. Analysis of tSNR confirmed improved data quality in the time domain that facilitates time-series analysis for task-fMRI. While NORDIC provided excellent performance and is a parameter free approach, it is computationally demanding and requires phase and magnitude data for denoising. Our KWIA approach also improved noise characteristics as compared to the standard reconstruction and might be an easy to implement and computationally fast and straightforward alternative to NORDIC for high resolution fMRI data.Acknowledgements
This study was supported by NIH grant U01EB029823. We thank Dr. Steen Moeller for sharing NORDIC code.References
Cortese S, Kelly C, Chabernaud C, et al. Toward systems neuroscience of ADHD: a meta-analysis of 55 fMRI studies. Am J Psychiatry. Oct 2012;169(10):1038-55. doi:10.1176/appi.ajp.2012.111015212.
Mwansisya TE, Hu A, Li Y, et al. Task and resting-state fMRI studies in first-episode schizophrenia: A systematic review. Schizophr Res. 11 2017;189:9-18. doi:10.1016/j.schres.2017.02.0263.
Lee MH, Smyser CD, Shimony JS. Resting-state fMRI: a review of methods and clinical applications. AJNR Am J Neuroradiol. Oct 2013;34(10):1866-72. doi:10.3174/ajnr.A32634.
Moeller S, Pisharady PK, Ramanna S, et al. NOise reduction with DIstribution Corrected (NORDIC) PCA in dMRI with complex-valued parameter-free locally low-rank processing. Neuroimage. 02 01 2021;226:117539. doi:10.1016/j.neuroimage.2020.1175395.
Zhao C, Shao X, Yan L, Wang DJJ. k-space weighted image average (KWIA) for ASL-based dynamic MR angiography and perfusion imaging. Magn Reson Imaging. 02 2022;86:94-106. doi:10.1016/j.mri.2021.11.0176.
Griswold MA, Jakob PM, Heidemann RM, et al. Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med. Jun 2002;47(6):1202-10. doi:10.1002/mrm.101717.
Vizioli L, Moeller S, Dowdle L, et al. Lowering the thermal noise barrier in functional brain mapping with magnetic resonance imaging. Nat Commun. 08 30 2021;12(1):5181. doi:10.1038/s41467-021-25431-8
Figures