2540
Comparison of test–retest reliability of structural, resting state functional, and diffusion tensor magnetic resonance imaging
Di Hu1, Yanqiu Lv1, Dandan Zheng2, Geli Hu2, Peng Sun2, and Yun Peng1
1The Department of Radiology,, Beijing Children's Hospital, Capital Medical University, National Center for Children's Health, Beijing, China, 2Clinical & Technical Support, Philips Healthcare, Beijing, China
1The Department of Radiology,, Beijing Children's Hospital, Capital Medical University, National Center for Children's Health, Beijing, China, 2Clinical & Technical Support, Philips Healthcare, Beijing, China
Synopsis
Keywords: Data Analysis, Reproductive
We measured imaging reproducibility in structural, resting state fMRI and diffusion tensor scans across different time points and scanners in healthy volunteers. First, we assessed structural imaging variability by calculating volume for seven subcortical structures. Second, we evaluated across-scanner and across-time reliability of rsfMRI by assessing temporal signal-to-noise ratio of five networks. Finally, we assessed variability in diffusion metrics across scanners and time points. Our results provide statistical validations for longitudinal work on multiple systems, especially for structural study. The influence of different equipment in rsfMRI and DTI related research may be considered, especially DMN and GCC analysis involved.Introduction:
Longitudinal MRI studies are becoming increasingly important to study structural and functional changes in the brain following pathophysiological conditions or therapeutic treatment. As the long time duration and multi-center approach were involved more and more in clinical research, the combining images acquired at different time points and different scanners require comparable and stable MR imaging measurements over time and across sites. In this study, we wanted to go beyond this, by measuring imaging reproducibility in structural, resting state fMRI and diffusion tensor scans across different time points and scanners in healthy volunteers. First, we assessed structural imaging variability across different scanners and time points by calculating volume for seven subcortical structures which are of particular interest in neurodegenerative diseases. Second, we evaluated across-scanner and across-time reliability of rsfMRI by assessing temporal signal-to-noise ratio (tSNR) and the generation of five networks in healthy volunteers. Finally, we assessed variability in diffusion metrics across scanners and time points.Materials and Methods:
Subjects were six healthy adults (3 males and 3 females) with an age range of 23 – 38 years old. The scan is carried out on different scanners on the same day and on the same scanner every other week to evaluate the consistency of different equipment and the stability of the same equipment. For each subject the scans on both scanners were conducted at approximately the same time of day. Subjects were advised to maintain the same caffeine intake on scan days and same sleep schedule the nights before. They were advised not to exercise on the day prior to scanning and on the day of scanning before the scan. MRI scans were performed on two 3 T MRI scanner: Philips Achieva 3.0T with an 8-channel head coil and Philips Ingenia CX 3.0T with a 15 channel dStream T/R head coil. High-resolution 3D-T1 weighted images were acquired following the ADNI protocol developed for multi-center intervendor acquisitions [1]. rsfMRI data were acquired using whole brain T2*-weighted gradient-echo EPI, sensitive to blood oxygenation level-dependent (BOLD) contrast. Diffusion-weighted imaging was performed with a spin-echo echo-planar-imaging (SE-EPI) sequence using 64 different diffusion directions and a b value of 1000 s/mm2. For 3D T1, the volume for the following 7 ROIs (Thalamus (right), Putamen (right), Hippocampus (right), Amygdala (right), brain stem, white matter (WM) and cerebrospinal fluid (CSF)) were estimated. For rsfMRI, tSNR within ROIs were estimated as the ratio of mean signal from all the voxels within the ROIs divided by the standard deviation across time [2]. We analyses the tSNR within five functional networks, the gray matter (GM), default-mode (DMN), ventral attention net (VAN), visual net(VIS) and the somatomotor(SM) network. For DTI, the mean and standard deviation of FA value across different conditions for genu of corpus callosum(GCC), body of corpus callosum(BCC), splenium of corpus callosum(SCC) and the total were calculated. One-way ANOVA was used to test group mean differences. ICC was used to measure test-retest reliability. A statistical threshold of p<0.05 was considered significant.Results:
For 3D T1, the ICC values of the volume of the seven ROIs indicating that there are no significant differences between the two time points in two scanners compared to the differences between the subjects, as shown in Table 1. And the intrascanner coefficients in different brain regions showed excellent reproducibility, especially in the right amygdala, as shown in Table 2. For rsfMRI, the mean and standard error of tSNR and the ICC across the two time points for the five networks were shown in Figure 1. And the intrascanner coefficients in different networks were shown in Table 3. The reproducibility of tSNR were from moderate to good, with the worst performance in DMN. For DTI, the ICC of FA values across the two time points for the corpus callosum regions were demonstrated in Table 4. It illustrated that the time reproducibility of FA values in corpus callosum was good in both scanners, with the ICC value of GCC was lower. Besides, the intrascanner coefficients of FA in corpus callosum, BCC and SCC was 0.92,0.98 and 0.81, respectively. However, that value in GCC was 0.45.Discussion and Conclusion:
This study reported a group of metrics used to quantify the reproducibility for standard neuroimaging data (structural, BOLD and DTI) collected across two identical 3.0 T scanners and at two time points. Our results show the metrics proposed are highly reproducible for volumetric data either between same scanner at two time points or between the two different scanners. The rsfMRI showed stable temporal SNR of subject’s networks across time points except DMN. And the reproducibility was moderate cross scanners. There was low variation of FA measures between scans for both intra- and inter-scanner rescanning in corpus callosum. While, when further analysis, the reproducibility of FA in GCC was poor across scanners which may be affected by the registration and segmentation. These findings provide statistical validations for longitudinal work on multiple systems, especially for structural study. The influence of different equipment in rsfMRI and DTI related research may be considered when analyzing data, especially DMN and GCC analysis involved.Acknowledgements
No acknowledgement found.References
[1] Jack CR Jr, Bernstein MA, Fox NC et al. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J Magn Reson Imaging ,2008,27:685–691. https://doi.org/10.1002/jmri.21049 [2] Parrish TB, Gitelman DR, LaBar KS, Mesulam MM. Impact of signal-to-noise on functional MRI. Magn Reson Med, 2000, 44: 925–9Figures
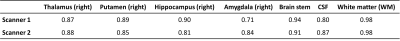
Table 1. Comparison of the time consistency of the volumes
in different brain regions measured by two scanners (ICC)

Table 2. Comparison of the inter scanner consistency of the volumes
in different brain regions(ICC)
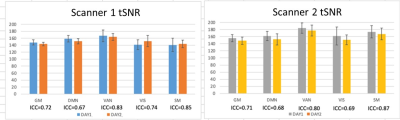
Figure 1. The mean and standard error of
tSNR and the ICC across the two time points for the five networks.

Table 3. Comparison of the inter scanner consistency of the
tSNR in five
functional networks (ICC)
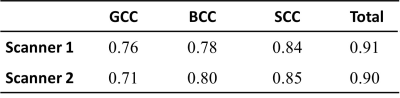
Table 4. Comparison of the time consistency of the
FA values in
different brain regions measured by two scanners (ICC)
DOI: https://doi.org/10.58530/2023/2540