2524
Mesoscale myelin-water fraction and T1/T2/PD mapping using optimized 3D ViSTa-MR Fingerprinting1Department of Radiology, Stanford University, Stanford, CA, United States, 2Department of Electrical Engineering, Stanford University, Stanford, CA, United States, 3Department of Electrical Engineering and Computer Science, Massachusetts Institute of Technology, Cambridge, MA, United States, 4Center for Brain Imaging Science and Technology, College of Biomedical Engineering & Instrument Science, Zhejiang University, Hangzhou, China, 5Department of Psychology, Stanford University, Stanford, CA, United States, 6Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital and Harvard Medical School, Charlestown, MA, United States, 7Techna Institute, University Health Network, Toronto, ON, Canada, 8Department of Imaging Sciences, University of Rochester, Rochester, NY, United States, 9Stanford Center for Cognitive and Neurobiological Imaging, Stanford University, Stanford, CA, United States
Synopsis
Keywords: Normal development, Microstructure
In this work, we developed an optimized ViSTa-MRF method, which combined Visualization of Short Transverse relaxation time component (ViSTa) technique with MR Fingerprinting (MRF), to achieve high-fidelity whole-brain myelin-water fraction (MWF) and T1/T2/PD mapping at sub-millimeter isotropic resolution. To achieve high image quality, fast acquisition, and memory-efficient reconstruction, the proposed ViSTa-MRF sequence leverages a CRLB-optimized flip-angle (FA) protocol, SNR-efficient 3D spiral-projection sampling scheme and a GPU-based subspace reconstruction. We also applied the proposed method to enable high-resolution assessment of MWF/T1/T2 for infant brain development as well as for post-mortem brain sample.Introduction
Myelin-Water Fraction (MWF)-mapping has shown great potential in characterizing brain’s myelination processes(1). To improve MWF-mapping, the ViSTa technique(2) that employed a specifically configured double-inversion-recovery was proposed to suppress the long T1-component for direct visualization of short-T1 myelin-water components. Our previous work(3) incorporated ViSTa into 3D-MR fingerprinting with 3D-tiny-golden-angle-shuffling(TGAS) acquisition and stochastic subspace reconstruction, to improve the SNR of the ViSTa and accelerate MWF-mapping, which enables whole-brain 0.66mm3 MWF and T1/T2/PD maps in 22.8 minutes.Building on the previous work, we developed approaches to further improve the fidelity of ViSTa-MRF: (i) We utilize the Cramér-Rao lower bound (CRLB) sequence parameters optimization(4,5) to increase the signal-to-noise-ratio(SNR), (ii) To enable fast subspace reconstruction for large mesoscopic-resolution ViSTa-MRF data, a novel polynomial preconditioned FISTA reconstruction with Pipe-Menon density-compensation was implemented with careful memory allocation to minimize memory footprint(6). We demonstrate that the proposed method achieves high-fidelity whole-brain MWF/T1/T2/PD maps at 0.66-mm-isotropic resolution in 15.8 minutes and post-mortem brain at 0.50-mm-isotropic resolution on a 3T clinical scanner. Furthermore, we propose a 5-minute whole-brain 1mm-iso ViSTa-MRF protocol to quantitively investigate brain development in early childhood.
Methods
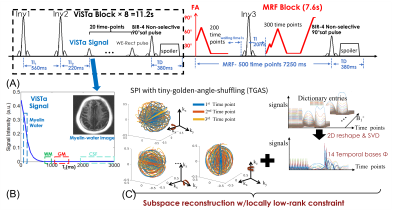
Pulse sequence: Figure1(A) shows the diagram of the ViSTa-MRF sequence, where each acquisition-group consists of eight ViSTa-blocks and one MRF-block. In each ViSTa-block, a double-inversion-recovery is performed, with the first subsequent signal time-point labeled the “ViSTa signal”. Through extended-phase-graph(EPG) simulation(7), Fig.1(B) shows that short-T1 signals including the myelin-water signal are preserved, while the long-T1 signals are suppressed, which enables direct myelin-water imaging.Reconstruction: Figure 1(C) shows the flowchart of sampling trajectories and subspace reconstruction with locally low-rank constraint. The ViSTa-MRF dictionary was generated using EPG, and the first 14 principal-components were selected as the temporal basesΦ. The ViSTa-MRF time-series x is then expressed as x=Φc, where c are the temporal coefficient maps. To reconstruct the whole-brain mesoscale resolution data with enough temporal-subspace bases, we implemented a memory-efficient version of polynomial-preconditioned FISTA algorithm with density-compensation in SigPy(8), where the 14-bases subspace reconstruction can be processed on a GPU with 24GB VRAM. The reconstructed c are then used to estimate T1/T2/PD maps. In addition, the quantitative MWF-map is derived by dividing the first time-point(ViSTa image) by the PD map.
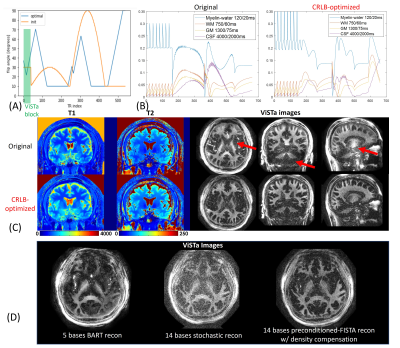
Protocol optimization: The CRLB method was adopted to optimize the FA pattern of the ViSTa-MRF sequence(Fig.2(A)) to improve the accuracy of the estimation of myelin-water, white-matter, and gray-matter. Figure2(B) shows original and CRLB-optimized ViSTa-MRF signal-curves for different tissue-types.
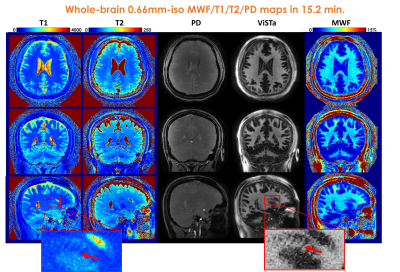
Experiments: All experiments were performed on a 3T GE UHP scanner with the approval of the institutional review board(IRB). 3D spiral-projection-imaging(9) was used for ViSTa-MRF acquisition(Fig.1(C)): FOV:220×220×220mm3, TR/TE=12/1.8ms with a 6.8ms spiral-readout. Forty-eight acquisition-groups were acquired for the 0.66-mm case. This resulted in a scan time of 19s×48=15.2minutes for 0.66mm-iso datasets.
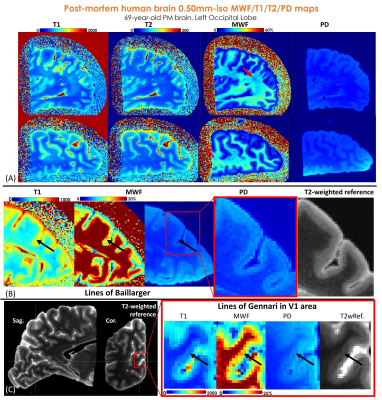
To validate the image quality of the proposed method, a left occipital lobe sample from a 69-year-old post-mortem brain was acquired with ViSTa-MRF at 0.50mm-isotropic resolution, FOV:160×160×160mm3, 180 acquisition-groups were acquired with a total acquisition time of 19s×180=57minutes. For this ex-vivo scan, a lower acceleration rate than feasible was used to ensure high SNR.
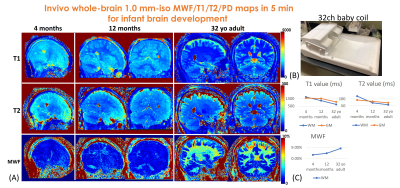
To quantitatively investigate infant brain development using MWF/T1/T2 maps, a 5-minute 1.0mm-iso whole-brain ViSTa-MRF protocol was utilized to acquire data on a 4-month and a 12-month infant. A custom-built tight-fitting 32-channel baby coil(Fig.5(B)) is used to acquire datasets for improved SNR. FOV:220×220×220mm3, 16 acquisition-groups were acquired, which resulted in 19s×16=5.0minutes.
To correct for B1+ inhomogeneity-related bias in ViSTa-MRF, a FOV-matched low-resolution Bloch-Siegert B1+ Mapping(11) was obtained.
Results
Figure2(C) shows the T1/T2/MWF maps using the original- and the CRLB-optimized FAs. The red arrows indicate the CRLB-optimized results achieve higher SNR in MWF maps. Figure2(D) demonstrates the superiority of the preconditioned-FISTA method with density-compensation compared to the memory-intensive 5-base BART reconstruction(12) and our previous 14-base stochastic-reconstruction(3). For the 1mm-iso whole-brain data, the preconditioned-FISTA subspace reconstruction uses ~5GB-VRAM and the total reconstruction time is 50minutes.Figure3 shows whole-brain 0.66mm-iso T1/T2/PD/ViSTa and MWF maps in three orthogonal views, where the red arrows in the zoom-in figures indicate the ability to visualize subtle brain structures such as the caudate nucleus in the dataset.
Figure4 shows the 0.50mm-iso ViSTa-MRF results of the post-mortem brain sample. As the red arrow indicated in Fig.4(A), the “dark-dots” in MWF imply the de-myelination in this region. Figure4(B) and (C) show decreased T1 & PD and increased MWF values (indicated in black arrows) in the lines of Baillarger in lateral occipital(13) and Gennari in V1 region, respectively, reflecting the high myelination in Layer IV of the cortex, which is consistent with the high-resolution T2-weighted images.
Figure5(A) shows the estimated 1mm-iso whole-brain T1/T2/MWF maps of 4-month, and 12-month babies and a reference adult. As shown in Fig.5(B), both T1 and T2 values of whole-brain white-matter and gray-matter decrease while the MWF of white-matter increases with brain development, indicating brain dynamic process of dendritic and axonal growth, and myelination(14,15).
Discussion and conclusion
In this work, we developed a 3D ViSTa-MRF sequence with CRLB-optimized FAs and a memory-efficient subspace reconstruction to achieve whole-brain mesoscale MWF and T1/T2/PD mapping in a single scan. Our preliminary results of two infant scans demonstrate the feasibility of using this technology for investigating brain development in early childhood.Acknowledgements
The authors would like to thank Dr. Jonathan R Polimeni (Massachusetts General Hospital) for the insightful discussions. The assistance by Clara Maria Bacmeister and Bella Fascendini (Stanford University) in preparing the experiments is also appreciated.
This study is supported in part by GE Healthcare and NIH fundings: R01-EB020613, R01-EB019437, R01-MH116173, P41EB030006, and U01-EB025162.
References
1. Piredda GF, Hilbert T, Thiran JP, Kober T. Probing myelin content of the human brain with MRI: A review. Magn. Reson. Med. 2021;85:627–652 doi: 10.1002/mrm.28509.
2. Oh SH, Bilello M, Schindler M, Markowitz CE, Detre JA, Lee J. Direct visualization of short transverse relaxation time component (ViSTa). Neuroimage 2013;83:485–492 doi: 10.1016/j.neuroimage.2013.06.047.
3. Liao C, Cao X, Iyer SS, et al. Mesoscale myelin-water fraction and T1/T2/PD mapping through optimized 3D ViSTa-MRF and stochastic reconstruction with preconditioning. In: ISMRM. London; 2022. p. 0365.
4. Zhao B, Haldar JP, Setsompop K, Wald LL. Optimal experiment design for magnetic resonance fingerprinting. In: 2016 38th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC). ; 2016. pp. 453–456. doi: 10.1109/EMBC.2016.7590737.
5. Lee PK, Watkins LE, Anderson TI, Buonincontri G, Hargreaves BA. Flexible and efficient optimization of quantitative sequences using automatic differentiation of Bloch simulations. Magn. Reson. Med. 2019;82:1438–1451 doi: 10.1002/mrm.27832.
6. Iyer SS, Ong F, Cao X, et al. Polynomial Preconditioners for Regularized Linear Inverse Problems. 2022 doi: 10.48550/arxiv.2204.10252.
7. Weigel M. Extended phase graphs: Dephasing, RF pulses, and echoes - Pure and simple. J. Magn. Reson. Imaging 2015;41:266–295 doi: 10.1002/jmri.24619.
8. Ong F, Lustig M. SigPy: a python package for high performance iterative reconstruction. In: ISMRM. ; 2019.
9. Cao X, Liao C, Srinivasan Iyer S, et al. Optimized multi-axis spiral projection MR fingerprinting with subspace reconstruction for rapid whole-brain high-isotropic-resolution quantitative imaging.
10. Jenkinson M, Bannister P, Brady M, Smith S. Improved Optimization for the Robust and Accurate Linear Registration and Motion Correction of Brain Images. Neuroimage 2002;17:825–841 doi: 10.1006/nimg.2002.1132.
11. Sacolick LI, Wiesinger F, Hancu I, Vogel MW. B1 mapping by Bloch-Siegert shift. Magn. Reson. Med. 2010;63:1315–1322 doi: 10.1002/MRM.22357.
12. Uecker M, Ong F, Tamir J, … DB-PIS, 2015 U. Berkeley advanced reconstruction toolbox. ISMRM 2015:2486.
13. Fracasso A, Van Veluw SJ, Visser F, et al. Lines of Baillarger in vivo and ex vivo: Myelin contrast across lamina at 7 T MRI and histology. Neuroimage 2016;133:163–175 doi: 10.1016/j.neuroimage.2016.02.072.
14. Chen Y, Chen MH, Baluyot KR, Potts TM, Jimenez J, Lin W. MR fingerprinting enables quantitative measures of brain tissue relaxation times and myelin water fraction in the first five years of life. Neuroimage 2019;186:782–793 doi: 10.1016/j.neuroimage.2018.11.038.
15. Huttenlocher PR. Morphometric study of human cerebral cortex development. Neuropsychologia 1990;28:517–527 doi: 10.1016/0028-3932(90)90031-I.
Figures