2500
Combined new molecular signature and radiological signature for risk stratification in IDH wild-type lower-grade glioma1Shanxi Medical University, Taiyuan, China
Synopsis
Keywords: Tumors, Cancer, Iower-grade glioma
When radiological signature was combined with new molecular signature, higher performance was achieved in predicting the prognosis of IDH wild-type lower-grade glioma patients. The combined model showed great risk stratification ability, which improving survival prediction in patients with IDH wild-type lower-grade glioma.INTRODUCTION
According to the 2021 World Health Organization (WHO) classification of central nervous system tumors, molecular diagnosis plays an important role in the diagnosis of isocitrate dehydrogenase-wildtype (IDHwt) diffuse gliomas. IDHwt lower-grade gliomas (LGGs) with one or more of the molecular signature, which include: epidermal growth factor receptor (EGFR) amplification, telomerase reverse transcriptase promoter (TERTp) mutations and chromosome 10 (whole chromosome, 10p or 10q) deletion and gain of chromosome 7 (whole chromosome, 7p or 7q), are expected to be more aggressive and have a worse prognosis.As a non-invasive examination method, neuroimaging not only plays an important role in the diagnosis and differential diagnosis of tumors, but also provides imaging information of tumors, which is also useful in molecular diagnosis and prognosis prediction of tumors. However, previous studies were mostly based on the 2016 WHO tumor classification criteria, and there were few studies on combining the radiological signature and molecular signature of IDHwt LGGs to predict the prognosis of IDHwt LGGs. Therefore, if radiological signature can be combined with molecular signature, the survival prediction ability of IDH wild-type LGGs may be improved.
The purpose of this study was to combine new molecular signature with radiological signature to construct an IDHwt LGGs model with better predictive ability.
METHODS
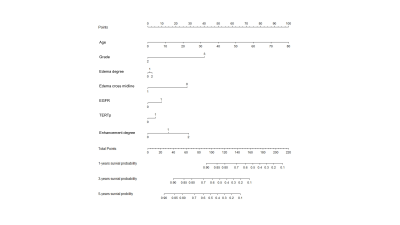
A total of 93 patients were collected and divided into training cohorts and validation cohorts using computer-generated random numbers in a 7:3 ratio stratified sampling. Random survival forest model was used to establish clinical gene model, radiological model and combined model. Evaluate the three models using the concordance index (C-index), the integrated Brier score (IBS) and the integrated area under the receiver operating characteristic curve (iAUC). All patients were divided into high-risk or low-risk groups according to the best model. Survival was calculated using the Kaplan-Meier method, and differences in overall survival between groups were compared using the log-rank test.RESULTS
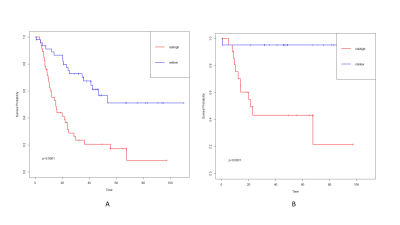
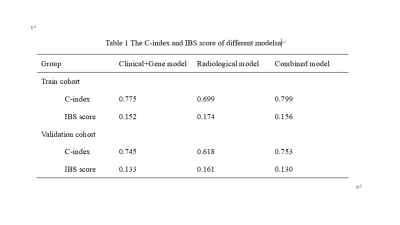
Combined model had the best performance than the other two models (C-index: 0.799 vs 0.775 and 0.699; IBSl:0.156 vs 0.152 and 0.174). The combined model had a higher iAUC value, the difference was statistically significant (difference to clinical gene model: 0.021[95%CI:0.005,0.059], P=0.035 and difference to radiological model:0.091[95%CI:0.031,0.157], P=0.023). Risk stratification of patients can be accurately performed in train cohort and validation cohort (both P<0.0001)DISCUSSION
When radiological signature was combined with new molecular signature, higher performance was achieved in predicting the prognosis of IDHwt LGGs patients. The combined model showed good risk stratification ability in both training and validation cohorts, which improving survival prediction in patients with IDHwt LGGs.Several previous studies have applied radiological features to predict survival in LGG patients. However, these studies lack the information of EGFR amplification and TERTp mutation status, which were key molecular markers for stratification of LGG prognosis with IDHwt according to the 2021 WHO classification. In this study, the combined model combining radiological and new molecular features exhibited higher diagnostic performance, suggesting that radiological features play an important role in improving the prognostic predictive power of IDHwt LGGs.
CONCLUSION
We build a combined model incorporating molecular and radiological signature that could well predict the prognosis of patients with IDHwt LGGs. Integrating molecular and radiological signature can improve survival prediction in patients with IDHwt LGGs.Acknowledgements
Acknowledgment
The authors want to thank the Shanxi Medical University, participants who took part in the study and data collection.
Ethical approval
The study was approved by the Ethics and Human Committees of Shanxi Medical University.
Consent to participate
All participants voluntarily participated and signed an informed consent form.
Consent for publication
All authors provide consent for publication.
Conflict of Interest
The authors report no conflict of interest.
Availability of data and material
The data for this study are not publicly available because the First Clinical Medical Hospital of Shanxi Medical University, the center from which the data were collected, does not agree to make the data publicly accessible. Further inquiry about data sharing maybe directed to Prof. Yan Tan, tanyan123456@sina.com.
References
1. Louis, D.N., et al., The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol, 2021. 23(8): p. 1231-1251.
2. Fujimoto, K., et al., TERT promoter mutation status is necessary and sufficient to diagnose IDH-wildtype diffuse astrocytic glioma with molecular features of glioblastoma. Acta Neuropathol, 2021. 142(2): p. 323-338.
3. Brat, D.J., et al., cIMPACT-NOW update 5: recommended grading criteria and terminologies for IDH-mutant astrocytomas. Acta Neuropathol, 2020. 139(3): p. 603-608.
4. Stichel, D., et al., Distribution of EGFR amplification, combined chromosome 7 gain and chromosome 10 loss, and TERT promoter mutation in brain tumors and their potential for the reclassification of IDHwt astrocytoma to glioblastoma. Acta Neuropathol, 2018. 136(5): p. 793-803.
5. Giannini, C. and F. Giangaspero, TERT promoter mutation: is it enough to call a WHO grade II astrocytoma IDH wild-type glioblastoma? Neuro Oncol, 2021. 23(6): p. 865-866.
6. Calabrese, E., et al., Combining radiomics and deep convolutional neural network features from preoperative MRI for predicting clinically relevant genetic biomarkers in glioblastoma. Neurooncol Adv, 2022. 4(1): p. vdac060.
7. Rathore, S., et al., Combining MRI and Histologic Imaging Features for Predicting Overall Survival in Patients with Glioma. Radiol Imaging Cancer, 2021. 3(4): p. e200108.
8. Sun, C., et al., Radiomics and Qualitative Features From Multiparametric MRI Predict Molecular Subtypes in Patients With Lower-Grade Glioma. Front Oncol, 2021. 11: p. 756828.
9. Tan, Y., et al., Whole-tumor radiomics analysis of DKI and DTI may improve the prediction of genotypes for astrocytomas: A preliminary study. Eur J Radiol, 2020. 124: p. 108785.
10. Tan, Y., et al., A radiomics nomogram may improve the prediction of IDH genotype for astrocytoma before surgery. Eur Radiol, 2019. 29(7): p. 3325-3337.
11. Park, C.J., et al., Radiomics risk score may be a potential imaging biomarker for predicting survival in isocitrate dehydrogenase wild-type lower-grade gliomas. Eur Radiol, 2020. 30(12): p. 6464-6474.
12. Wang, H. and L. Zhou, Random survival forest with space extensions for censored data. Artif Intell Med, 2017. 79: p. 52-61.
13. Bae, S., et al., Radiomic MRI Phenotyping of Glioblastoma: Improving Survival Prediction. Radiology, 2018. 289(3): p. 797-806.
14. Chen, J., et al., Peritumor Edema Serves as an Independent Predictive Factor of Recurrence Patterns and Recurrence-Free Survival for High-Grade Glioma. Comput Math Methods Med, 2022. 2022: p. 9547166.
15. Pei, W., et al., MRI-based random survival Forest model improves prediction of progression-free survival to induction chemotherapy plus concurrent Chemoradiotherapy in Locoregionally Advanced nasopharyngeal carcinoma. BMC Cancer, 2022. 22(1).
16. Adeoye, J., et al., Deep Learning Predicts the Malignant-Transformation-Free Survival of Oral Potentially Malignant Disorders. Cancers (Basel), 2021. 13(23).
17. Reps, J.M., P. Ryan, and P.R. Rijnbeek, Investigating the impact of development and internal validation design when training prognostic models using a retrospective cohort in big US observational healthcare data. BMJ Open, 2021. 11(12).
18. Takada, T., et al., Internal-external cross-validation helped to evaluate the generalizability of prediction models in large clustered datasets. Journal of Clinical Epidemiology, 2021. 137: p. 83-91.
19. Fukuma, R., et al., Prediction of IDH and TERT promoter mutations in low-grade glioma from magnetic resonance images using a convolutional neural network. Sci Rep, 2019. 9(1): p. 20311.
20. Berzero, G., et al., IDH-wildtype lower-grade diffuse gliomas: the importance of histological grade and molecular assessment for prognostic stratification. Neuro Oncol, 2021. 23(6): p. 955-966.
21. Olar, A., et al., IDH mutation status and role of WHO grade and mitotic index in overall survival in grade II-III diffuse gliomas. Acta Neuropathol, 2015. 129(4): p. 585-96.
22. Gittleman, H., A.E. Sloan, and J.S. Barnholtz-Sloan, An independently validated survival nomogram for lower-grade glioma. Neuro Oncol, 2020. 22(5): p. 665-674.
23. Komori, T., Updating the grading criteria for adult diffuse gliomas: beyond the WHO2016CNS classification. Brain Tumor Pathol, 2020. 37(1): p. 1-4.
24. Tesileanu, C.M.S., et al., Survival of diffuse astrocytic glioma, IDH1/2 wildtype, with molecular features of glioblastoma, WHO grade IV: a confirmation of the cIMPACT-NOW criteria. Neuro Oncol, 2020. 22(4): p. 515-523.
25. Wu, C.X., et al., Peritumoral edema on magnetic resonance imaging predicts a poor clinical outcome in malignant glioma. Oncol Lett, 2015. 10(5): p. 2769-2776.
26. Mummareddy, N., et al., Prognostic relevance of CSF and peri-tumoral edema volumes in glioblastoma. J Clin Neurosci, 2021. 84: p. 1-7.
27. Ning, L., et al., Correlations between Clinical Characteristics and Prognosis in Patients with Grade II Glioma. Evid Based Complement Alternat Med, 2021. 2021: p. 5873213.
28. Tunthanathip, T., S. Sangkhathat, and K. Kanjanapradit, Risk Factors Associated with Malignant Transformation of Astrocytoma: Competing Risk Regression Analysis. Asian J Neurosurg, 2022. 17(1): p. 3-10.
29. Zhao, M., et al., Quantitative analysis of permeability for glioma grading using dynamic contrast-enhanced magnetic resonance imaging. Oncol Lett, 2017. 14(5): p. 5418-5426.
30. Leroy, H.A., et al., High-field intraoperative MRI in glioma surgery: A prospective study with volumetric analysis of extent of resection and functional outcome. Neurochirurgie, 2018. 64(3): p. 155-160.
31. Malik, N., et al., MRI radiomics to differentiate between low grade glioma and glioblastoma peritumoral region. J Neurooncol, 2021. 155(2): p. 181-191.
32. Li, M., et al., Combining hyperintense FLAIR rim and radiological features in identifying IDH mutant 1p/19q non-codeleted lower-grade glioma. Eur Radiol, 2022. 32(6): p. 3869-3879.
33. Park, Y.W., et al., Adding radiomics to the 2021 WHO updates may improve prognostic prediction for current IDH-wildtype histological lower-grade gliomas with known EGFR amplification and TERT promoter mutation status. Eur Radiol, 2022.
34. Park, C.J., et al., MRI Features May Predict Molecular Features of Glioblastoma in Isocitrate Dehydrogenase Wild-Type Lower-Grade Gliomas. AJNR Am J Neuroradiol, 2021. 42(3): p. 448-456.
35. Choi, Y.S., et al., Machine learning and radiomic phenotyping of lower grade gliomas: improving survival prediction. Eur Radiol, 2020. 30(7): p. 3834-3842.
36. Gittleman, H., et al., An independently validated nomogram for individualized estimation of survival among patients with newly diagnosed glioblastoma: NRG Oncology RTOG 0525 and 0825. Neuro Oncol, 2017. 19(5): p. 669-677.
Figures
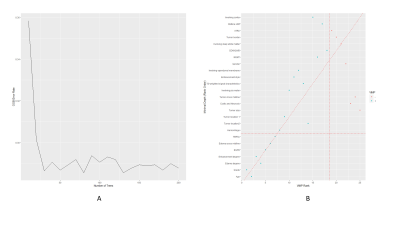
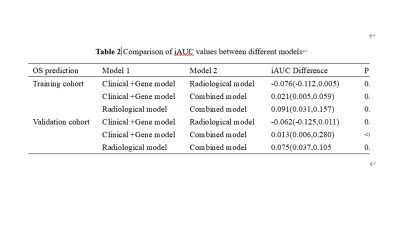
Comparison of iAUC values between different models
Note: The iAUC difference was calculated as (model 2 iAUC) 2 (model 1 iAUC). Data in parentheses are 95% confidence intervals.