2499
Convolutional neural network to predict IDH mutation status in glioma from 7T chemical exchange saturation transfer imaging1Department of neurosurgery, Huashan Hospital Fudan University, Shanghai, China, 2Department of radiology, Huashan Hospital Fudan University, Shanghai, China, 3MR Collaboration, Siemens Healthineers Ltd, Shanghai, China, 4School of Biomedical Engineering, ShanghaiTech University, shanghai, China
Synopsis
Keywords: Tumors, CEST & MT
Noninvasive prediction of isocitrate dehydrogenase (IDH) mutation status in glioma guides surgical strategies and individualized management. We explored the capability of preoperatively identifying IDH status by combining a 2D convolutional neural network (CNN) and amide proton transfer chemical exchange saturation transfer (APT-CEST) imaging. Five-fold cross-validation suggested the APT-CEST with the tumor shape information predicts IDH status optimally. The novel CNN model designed for 7T APT-CEST offers improved discriminatory accuracy in predicting the IDH status of glioma, holding great potential for facilitating decision-making in clinical practice.Introduction
Since the World Health Organization (WHO) 2016 classification of gliomas includes variations in underlying genetic and epigenetic alterations. More evidence has proved that these distinct genetic subtypes, such as a mutation in IDH or codeletion of 1p and 19q chromosomal arms, indicate drastic differences in overall survival and response to therapy [1-3]. Due to this increasing emphasis on molecular pathology as a diagnostic standard, genetic testing based on tissue acquired from surgery has been largely used to guide subsequent treatment. Given that genetic testing can be costly and time-consuming and there remain cases where resection is not recommended, an alternative, noninvasive approach for obtaining this crucial genetic information is desirable.Material and Methods
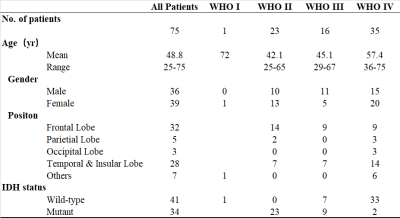
Seventy-five patients, who were newly diagnosed with a pathologically confirmed glioma, underwent MRI scanning on a 7T MRI system (MAGNETOM Terra; Siemens Healthineers, Erlangen, Germany). Among them, forty-one patients were diagnosed with IDH-wild type glioma, and thirty-four patients were diagnosed as IDH-mutant (Table 1). MRI images included amide proton transfer imaging via chemical exchange saturation transfer (APT-CEST) and T1 MP2RAGE; details of data acquisition can be found in [4].Data analysis
Experienced neurologists manually annotate the tumor regions in the T1 image. All images were co-registered to T1 images, cropped according to the tumor region bounding box, and resized into 100X100 pixels. Each pair of CEST, T1 and tumor annotated slices with IDH labels was considered as one sample, and a total of 4090 pairs were selected as our training and testing datasets [5,6].
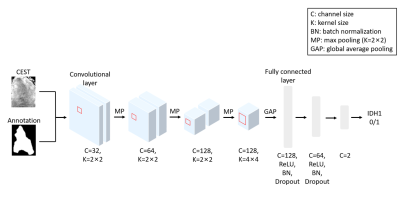
We designed a 2D convolutional neural network (CNN) to predict IDH-type. The preprocessed CEST and tumor annotation slices are combined as two channels and inputted into the CNN. Under this setting, our model considers the metabolism information from CEST and the (boundary) shape information of the tumor for IDH prediction. The architecture and the parameter settings are depicted in Figure 1. We train the CNN with training epoch =35, learning rate =0.01 for the first ten epochs and 0.001 for the 10th to 20th epochs, and 0.0001 for the remaining epochs, and batch size =32. Adam algorithm [7] was used to optimize the trainable parameters with a weight decay of 0.001. Since the sample size is slightly imbalanced in different groups, the weighted cross-entropy was employed as the loss function, with weights of 1 for IDH and 1.2 for mutant groups.
Validation scheme
Five-fold cross-validation was performed to assess the predictability of our method. The 4090 slices were randomly split into five equal folds, with four folds being the training set and the remaining one being the testing set. Five validations were performed, so each fold behaves as a testing set once. For predictions from each round, we compute four metrics from different aspects to evaluate the performance, i.e., the accuracy (ACC), sensitivity (SEN), specificity (SPE), and area under the receiver operating characteristic curve (AUC).
Statistical analysis
The means, standard deviation (STD), and 95% confidence interval (CI) of the performance metrics from cross-validation are calculated. The cross-validation comparisons are performed by paired t-test. In addition, we also integrate the predictions from each fold to compute the metrics on predictions for all the participants. The significance of this integrated result is estimated by a permutation test. The p-values are obtained by the probability of finding a metrics value that is larger than the value of the real metrics from CNN in the chance level distribution.
Results and discussion
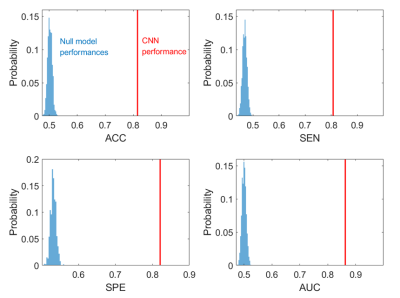
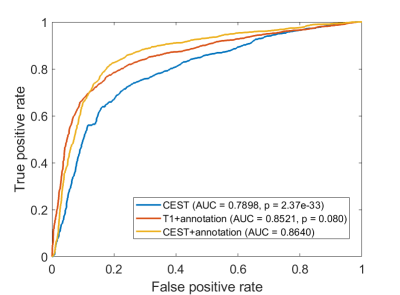
In the cross-validations, our method obtains ACC = 0.8149 (CI = [0.7908, 0.8390]), SEN = 0.8070 (CI = [0.7747, 0.8392]), SPE = 0.8208 (CI = [0.7807, 0.8609]), and AUC = 0.8629 (CI = [0.8391, 0.8866]). When integrating the predictions, the metrics values are ACC = 0.8149 (p<0.001), SEN = 0.8077 (p<0.001), SPE =0.8811 (p<0.001), and AUC = 0.8640 (p<0.001), all of which shows significance when comparing to the chance levels (Figure 2).In addition, we train the CNN using only the CEST images (Table 2, “CEST”). When using CEST only, the prediction performances drop to ACC = 0.7433 (CI = [0.7208, 0.7658]), SEN = 0.7860 (CI = [0.7034, 0.7219]), SPE = 0.7696 (CI = [0.7307, 0.8084]) and AUC = 0.7908 (CI = [0.7699, 0.8117]). These metrics are still higher than the chance level (see distributions in Figure 4). It suggests that CEST images can provide satisfactory predictability on the IDH1 mutation. On the other hand, all these metrics are significantly lower than the metrics from the “CEST+annotation” methods. The AUC of this setting is also significantly lower than the AUC based on CEST with annotation mask (p<0.001. Figure 3). It thus can indicate the importance of diagnostic shape information contained by the annotation masks.
Conclusion
The novel CNN model derived for 7T APT-CEST imaging offers improved discriminatory accuracy in predicting IDH status of glioma when compared to conventional T1 imaging, thus holding great potential for facilitating decision-making in clinical practice.Acknowledgements
No acknowledgement found.References
1. van den Bent MJ, Brandes AA, Taphoorn MJ, et al. Adjuvant procarbazine, lomustine, and vincristine chemotherapy in newly diagnosed anaplastic oligodendroglioma: long-term follow-up of EORTC brain tumor group study 26951. J Clin Oncol. 2013;31(3):344-350.
2.Hartmann C, Hentschel B, Wick W, et al. Patients with IDH1 wild type anaplastic astrocytomas exhibit worse prognosis than IDH1-mutated glioblastomas, and IDH1 mutation status accounts for the unfavorable prognostic effect of higher age: implications for classification of gliomas. Acta Neuropathol. 2010;120(6):707-718.
3.Iorgulescu JB, Sun C, Neff C, et al. Molecular biomarker-defined brain tumors: Epidemiology, validity, and completeness in the United States. Neuro Oncol. 2022;24(11):1989-2000.
4. Yuan, Yifan, et al. "Noninvasive Delineation of Glioma Infiltration with Combined 7T Chemical Exchange Saturation Transfer Imaging and MR Spectroscopy: A Diagnostic Accuracy Study." Metabolites 12.10 (2022): 901.
5.Fukuma, R., Yanagisawa, T., Kinoshita, M., Shinozaki, T., Arita, H., Kawaguchi, A., ... & Kishima, H. (2019). Prediction of IDH and TERT promoter mutations in low-grade glioma from magnetic resonance images using a convolutional neural network. Scientific reports, 9(1), 1-8.
6. Calabrese E, Rudie JD, Rauschecker AM, et al. Combining radiomics and deep convolutional neural network features from preoperative MRI for predicting clinically relevant genetic biomarkers in glioblastoma. Neurooncol Adv. 2022;4(1):vdac060.
7. Kingma, Diederik P., and Jimmy Ba. "Adam: A method for stochastic optimization." arXiv preprint arXiv:1412.6980 (2014).
Figures