2457
Deep-Learning-Based Knee Articular Cartilage Morphometrics
Yongcheng Yao1 and Weitian Chen1
1Department of Imaging and Interventional Radiology, The Chinese University of Hong Kong, Shatin, Hong Kong
1Department of Imaging and Interventional Radiology, The Chinese University of Hong Kong, Shatin, Hong Kong
Synopsis
Keywords: Cartilage, Cartilage, morphometrics
We proposed a deep-learning-based system for automatic knee articular cartilage morphometrics. It produces regional metrics including full-thickness cartilage loss (FCL), mean thickness, surface area, and volume. The proposed system comprises deep learning models and algorithms that work collaboratively. We have trained convolutional neural networks for tissue segmentation, template construction, and image registration. We designed modules and pipelines for cartilage thickness mapping, cartilage lesion quantification, and cartilage parcellation. Results shows superior accuracy of the thickness mapping method and robustness of the cartilage parcellation method. The proposed FCL estimation method filled the gap in automatic cartilage lesions quantification.Introduction
Knee articular cartilage morphometrics has been shown to be an effective tool for deriving imaging biomarkers for knee osteoarthritis (OA)1-8. Efforts have been made for the methodological development of cartilage morphometrics. Semiautomatic and semiquantitative methods have been proposed for the quantification or grading of cartilage lesions, thickness, surface area, and volume5, 9-19. Despite the successful applications of automatic tissue segmentation20-28, there is a lack of automatic methods for the quantification of cartilage lesions. To date, a fully automatic image analysis system for the quantification of normal cartilage anatomy and cartilage lesions has not been introduced. To overcome the drawbacks of semiautomatic and semiquantitative methods such as subjectivity and heavy time consumption, we developed a deep-learning-based system for automatic articular cartilage morphometrics. The proposed system takes a three-dimensional magnetic resonance image as input and produces regional metrics for each cartilage including full-thickness cartilage loss (FCL), mean thickness, surface area, and volume. The proposed system is part of the deep-learning-based platform, CartiMorph.The contributions of this work are summarized as follows: (1) we proposed a method for cartilage thickness mapping that is robust to lesions; (2) we proposed a method for FCL estimation; (3) we proposed a cartilage parcellation method for accurate regional quantification.
Methods
The proposed system (Fig. 1a) includes 3 deep-learning models for tissue segmentation, template construction (Fig. 1b left), and image registration (Fig. 1b right), respectively. Additionally, we designed modules for cartilage thickness mapping, FCL estimation (Fig. 1c), and cartilage parcellation. These models and modules work collaboratively. For example, the thickness mapping module relies on tissue segmentation; the FCL estimation module relies on tissue segmentation, template construction, and image registration; the cartilage parcellation module is based on FCL estimation.Overview. Given an input image $$$\boldsymbol{I}_i$$$, the network $$$\mathcal{F}_{\boldsymbol{\theta}_s}$$$ was trained to output a segmentation mask $$$\boldsymbol{S}_i$$$ via supervised learning. The thickness mapping module took the mask $$$\boldsymbol{S}_i$$$ and produced a cartilage thickness map through surface reconstruction, surface segmentation, and thickness measurement. The network $$$\mathcal{G}_{\boldsymbol{\theta}_t}$$$ was trained to build a representative template image $$$\boldsymbol{I}^t$$$ from a subset of masked, downsampled, and cropped images $$$\{\boldsymbol{I}_i^{low}\}_n$$$. A template segmentation $$$\boldsymbol{S}^t$$$ was constructed through manual labeling or template-to-image registration. The network $$$\mathcal{G}_{\boldsymbol{\theta}_u}$$$ was trained for image registration via unsupervised learning, from which a deformation field $$$\boldsymbol{\phi}_i$$$ was estimated and used for wrapping the template segmentation $$$\boldsymbol{S}^t$$$. The upsampled and wrapped template segmentation $$$(\boldsymbol{S}^t \circ \boldsymbol{\phi}_i)$$$, together with the segmentation mask $$$\boldsymbol{S}_i$$$, was used in FCL estimation. With the reconstructed surface from FCL estimation, each cartilage was divided into subregions which helped the regional metrics calculation.
Tissue Segmentation. We trained variants of nnU-Net29 and integrated the best model into the proposed system.
Template Construction. Inspired by a template learning model30 and the inverse-consistency constraint31, 32 for registration, we formulated the learning of template image $$$\widehat{\boldsymbol{I}^t}$$$ as the optimization (Fig. 1b left):
$$\underset{\boldsymbol{\theta}_v,\widehat{\boldsymbol{I}^t_i}}{\arg\,\min}\;\frac{1}{|\Omega|}\mathcal{L}_I(\boldsymbol{\theta}_v,\widehat{\boldsymbol{I}^t_i},\boldsymbol{I}^{low}_i),$$$$ \begin{eqnarray}\mathcal{L}_I = \quad &\lambda_1& \parallel \boldsymbol{I}^{low}_i - \widehat{\boldsymbol{I}^t_i} \circ \mathcal{I}(\mathcal{G}_{\boldsymbol{\theta}_v}^i(\widehat{\boldsymbol{I}^t_i},\boldsymbol{I}^{low}_i)) \parallel^2_2 \\&+& \lambda_2 \parallel \boldsymbol{I}^{low}_i \circ \mathcal{I}(-\mathcal{G}_{\boldsymbol{\theta}_v}^i (\widehat{\boldsymbol{I}^t_i},\boldsymbol{I}^{low}_i)) - \widehat{\boldsymbol{I}^t_i} \parallel^2_2 \\&+& \lambda_3 \parallel \mathcal{K}(\{ \mathcal{I}(-\mathcal{G}_{\boldsymbol{\theta}_v}^j (\widehat{\boldsymbol{I}^t_j},\boldsymbol{I}^{low}_j)) \}_{j \in N(i)}) \parallel^2_2 \\&+& \lambda_4 \parallel \nabla \mathcal{I}(\mathcal{G}_{\boldsymbol{\theta}_v}^i(\widehat{\boldsymbol{I}^t_i},\boldsymbol{I}^{low}_i)) \parallel^2_2 \end{eqnarray}.$$
Image Registration. We adopted VoxelMorph33 for deformable image registration. Given a random pair of moving and fixed images $$$\{\boldsymbol{I}_m, \boldsymbol{I}_f\}$$$, the network $$$\mathcal{G}_{\boldsymbol{\theta}_u}$$$ was trained by minimizing the loss function: $$\mathcal{L}(\boldsymbol{\theta}_u, \boldsymbol{I}_m, \boldsymbol{I}_f) = \mathcal{L}_{LNCC}(\boldsymbol{\theta}_u, \boldsymbol{I}_m, \boldsymbol{I}_f) + \frac{\lambda}{|\Omega|}\parallel\nabla\mathcal{G}_{\boldsymbol{\theta}_u}(\boldsymbol{I}_m,\boldsymbol{I}_f) \parallel^2_2 ,$$ where $$$\mathcal{L}_{LNCC}(\cdot)$$$ denotes local normalized cross-correlation loss and $$$\nabla$$$ is gradient operator.
Cartilage Thickness Mapping. We have developed a surface closing and restricted surface dilation algorithm for surface segmentation. We proposed a surface-normal-based thickness measurement method where normal vectors were estimated by singular value decomposition.
FCL Estimation. As shown in Fig. 1c, the FCL can be estimated from the proposed pipeline. We have developed complementary connectivity-based and curve-fitting-based surface-hold-filling algorithms for the reconstruction of FCL.
Cartilage Parcellation. We established a method for cartilage parcellation that is robust to FCL. A rule-based algorithm was developed and applied to the surface with reconstructed FCL.
Regional Quantification. We quantified the mean thickness over the reconstructed surface including the denuded area. We calculated FCL as the percentage of denuded area. Regional measurements of surface area and volume were also included.
Results
We used a subset of the public Osteoarthritis Initiative (OAI) dataset released by Zuse Institute Berlin (OAI-ZIB)34 in this work.Fig. 2 shows the example results for template construction, image registration, thickness mapping, and cartilage parcellation. Fig. 3 shows the performance of segmentation and registration networks on each cartilage and sample grouped by the Kellgren–Lawrence (KL) grade. We further evaluated the effectiveness of the segmentation model by comparing the regional metrics calculated from manual labels and those from model segmentation (Fig. 4).
Discussion
The proposed cartilage thickness mapping method shows superiority in peripheral areas and regions with thin cartilage compared with the nearest neighbor approach. Our FCL estimation method leverages the power of the unsupervised deformable registration model for template building. The novelty of the cartilage parcellation method resides in the combination of FCL estimation and the rule-based algorithm.Conclusion
Our FCL estimation method filled the gap in automatic cartilage lesions quantification. The proposed system, as a platform for imaging biomarkers extraction, may benefit keen-OA-related research.Acknowledgements
This work was supported by a grant from the Innovation and Technology Commission of the Hong Kong SAR [MRP/001/18X] and a grant from the Faculty Innovation Award of the Chinese University of Hong Kong.References
- Mosher, T.J., Zhang, Z., Reddy, R., Boudhar, S., Milestone, B.N., Morrison, W.B., Kwoh, C.K., Eckstein, F., Witschey, W.R., Borthakur, A., 2011. Knee articular cartilage damage in osteoarthritis: analysis of mr image biomarker reproducibility in acrin-pa 4001 multicenter trial. Radiology 258, 832.
- Hunter, D.J., Nevitt, M., Losina, E., Kraus, V., 2014. Biomarkers for osteoarthritis: current position and steps towards further validation. Best practice & research Clinical rheumatology 28, 61–71.
- Eckstein, F., Collins, J., Nevitt, M., Lynch, J., Kraus, V., Katz, J., Losina, E., Wirth, W., Guermazi, A., Roemer, F., et al., 2015. Brief report: cartilage thickness change as an imaging biomarker of knee osteoarthritis progression: data from the foundation for the national institutes of health osteoarthritis biomarkers consortium. Arthritis & rheumatology 67, 3184–3189.
- Collins, J.E., Losina, E., Nevitt, M.C., Roemer, F.W., Guermazi, A., Lynch, J.A., Katz, J.N., Kent Kwoh, C., Kraus, V.B., Hunter, D.J., 2016. Semiquantitative imaging biomarkers of knee osteoarthritis progression: data from the foundation for the national institutes of health osteoarthritis biomarkers consortium. Arthritis & rheumatology 68, 2422–2431.
- Guermazi, A., Hayashi, D., Roemer, F.W., Niu, J., Quinn, E.K., Crema, M.D., Nevitt, M.C., Torner, J., Lewis, C.E., Felson, D.T., 2017. Brief report: partial-and full-thickness focal cartilage defects contribute equally to development of new cartilage damage in knee osteoarthritis: the multicenter osteoarthritis study. Arthritis & rheumatology 69, 560–564.
- Wirth, W., Hunter, D., Nevitt, M., Sharma, L., Kwoh, C., Ladel, C., Eckstein, F., 2017. Predictive and concurrent validity of cartilage thickness change as a marker of knee osteoarthritis progression: data from the osteoarthritis initiative. Osteoarthritis and cartilage 25, 2063–2071.
- Everhart, J.S., Abouljoud, M.M., Kirven, J.C., Flanigan, D.C., 2019. Full-thickness cartilage defects are important independent predictive factors for progression to total knee arthroplasty in older adults with minimal to moderate osteoarthritis: data from the osteoarthritis initiative. JBJS 101, 56–63.
- Hunter, D.J., Deveza, L.A., Collins, J.E., Losina, E., Katz, J.N., Nevitt, M.C., Lynch, J.A., Roemer, F.W., Guermazi, A., Bowes, M.A., et al., 2022. Multivariable modeling of biomarker data from the phase i foundation for the national institutes of health osteoarthritis biomarkers consortium. Arthritis Care & Research.
- Cohen, Z.A., Mccarthy, D.M., Kwak, S.D., Legrand, P., Fogarasi, F., Ciaccio, E.J., Ateshian, G.A., 1999. Knee cartilage topography, thickness, and contact areas from mri: in-vitro calibration and in-vivo measurements. Osteoarthritis and cartilage 7, 95–109.
- Stammberger, T., Eckstein, F., Englmeier, K.H., Reiser, M., 1999. Determination of 3d cartilage thickness data from mr imaging: computational method and reproducibility in the living. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 41, 529–536.
- Hohe, J., Ateshian, G., Reiser, M., Englmeier, K.H., Eckstein, F., 2002. Surface size, curvature analysis, and assessment of knee joint incongruity with mri in vivo. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine 47, 554–561.
- Kauffmann, C., Gravel, P., Godbout, B., Gravel, A., Beaudoin, G., Raynauld, J.P., Martel-Pelletier, J., Pelletier, J.P., de Guise, J.A., 2003. Computer-aided method for quantification of cartilage thickness and volume changes using mri: validation study using a synthetic model. IEEE transactions on Biomedical Engineering 50, 978–988.
- Eckstein, F., Cicuttini, F., Raynauld, J.P., Waterton, J.C., Peterfy, C., 2006b.Magnetic resonance imaging (mri) of articular cartilage in knee osteoarthritis (oa): morphological assessment. Osteoarthritis and cartilage 14, 46–75.
- Carballido-Gamio, J., Bauer, J.S., Stahl, R., Lee, K.Y., Krause, S., Link, T.M., Majumdar, S., 2008. Inter-subject comparison of mri knee cartilage thickness. Medical image analysis 12, 120–135.
- Wirth, W., Eckstein, F., 2008. A technique for regional analysis of femorotibial cartilage thickness based on quantitative magnetic resonance imaging. IEEE transactions on medical imaging 27, 737–744.
- Williams, T.G., Holmes, A.P., Waterton, J.C., Maciewicz, R.A., Hutchinson, C.E., Moots, R.J., Nash, A.F., Taylor, C.J., 2010. Anatomically corresponded regional analysis of cartilage in asymptomatic and osteoarthritic knees by statistical shape modelling of the bone. IEEE transactions on medical imaging 29, 1541–1559.
- Maerz, T., Newton, M., Matthew, H., Baker, K., 2016. Surface roughness and thickness analysis of contrast-enhanced articular cartilage using mesh parameterization. Osteoarthritis and cartilage 24, 290–298.
- Favre, J., Erhart-Hledik, J.C., Blazek, K., Fasel, B., Gold, G.E., Andriacchi, T.P., 2017. Anatomically standardized maps reveal distinct patterns of cartilage thickness with increasing severity of medial compartment knee osteoarthritis. Journal of orthopaedic research 35, 2442–2451.
- Dório, M., Hunter, D., Collins, J., Asher, R., Eckstein, F., Guermazi, A., Roe- mer, F., Deveza, L., 2020. Association of baseline and change in tibial and femoral cartilage thickness and development of widespread full-thickness cartilage loss in knee osteoarthritis–data from the osteoarthritis initiative. Osteoarthritis and Cartilage 28, 811–818.
- Prasoon, A., Petersen, K., Igel, C., Lauze, F., Dam, E., Nielsen, M., 2013. Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network, in: International conference on medical image computing and computer-assisted intervention, Springer. pp. 246–253.
- Raj, A., Vishwanathan, S., Ajani, B., Krishnan, K., Agarwal, H., 2018. Automatic knee cartilage segmentation using fully volumetric convolutional neural networks for evaluation of osteoarthritis, in: 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), IEEE. pp. 851–854.
- Liu, F., Zhou, Z., Jang, H., Samsonov, A., Zhao, G., Kijowski, R., 2018. Deep convolutional neural network and 3d deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging. Magnetic resonance in medicine 79, 2379–2391.
- Zhou, Z., Zhao, G., Kijowski, R., Liu, F., 2018. Deep convolutional neural network for segmentation of knee joint anatomy. Magnetic resonance in medicine 80, 2759–2770.
- Ambellan, F., Tack, A., Ehlke, M., Zachow, S., 2019. Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: Data from the osteoarthritis initiative. Medical image analysis 52, 109–118.
- Tan, C., Yan, Z., Zhang, S., Li, K., Metaxas, D.N., 2019. Collaborative multi-agent learning for mr knee articular cartilage segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 282–290.
- Xu, Z., Niethammer, M., 2019. Deepatlas: Joint semi-supervised learning of image registration and segmentation, in: International Conference on Medical Image Computing and Computer-Assisted Intervention, Springer. pp. 420–429.
- Gaj, S., Yang, M., Nakamura, K., Li, X., 2020. Automated cartilage and meniscus segmentation of knee mri with conditional generative adversarial networks. Magnetic resonance in medicine 84, 437–449.
- Khan, S., Azam, B., Yao, Y., Chen, W., 2022. Deep collaborative network with alpha matte for precise knee tissue segmentation from mri. Computer Methods and Programs in Biomedicine 222, 106963.
- Isensee, F., Jaeger, P.F., Kohl, S.A., Petersen, J., Maier-Hein, K.H., 2021. nnu-net: a self-configuring method for deep learning-based biomedical image segmentation. Nature methods 18, 203–211.
- Dalca, A., Rakic, M., Guttag, J., Sabuncu, M., 2019a. Learning conditional deformable templates with convolutional networks. Advances in neural information processing systems 32.
- Zhang, J., 2018. Inverse-consistent deep networks for unsupervised deformable image registration. arXiv preprint arXiv:1809.03443.
- Mok, T.C., Chung, A., 2020. Fast symmetric diffeomorphic image registration with convolutional neural networks, in: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pp. 4644–4653.
- Balakrishnan, G., Zhao, A., Sabuncu, M.R., Guttag, J., Dalca, A.V., 2019. Voxelmorph: a learning framework for deformable medical image registration. IEEE transactions on medical imaging 38, 1788–1800.
- Ambellan F, Tack A, Ehlke M, Zachow S. Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: Data from the Osteoarthritis Initiative. Medical image analysis. 2019 Feb 1;52:109-18.
Figures
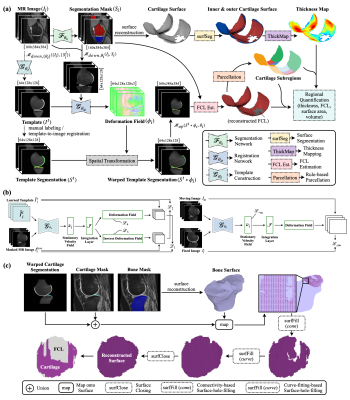
Figure 1. Methodology of the proposed system. (a) Overview of the proposed system. (b) The architectures of the template construction network (left) and the image registration network (right). (c) Pipeline of the proposed FCL estimation method.
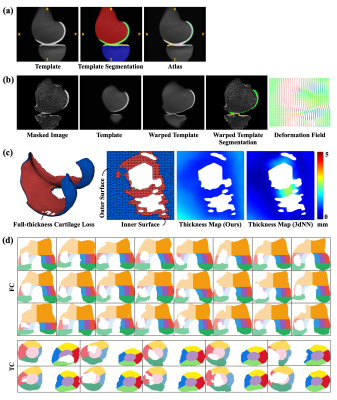
Figure 2. Example results from the proposed system. (a) The template image learned from the template construction network, the segmentation mask for the template image, and the 20-region atlas for the template image constructed by the proposed cartilage parcellation algorithm. (b) Results from template-to-image registration. (c) Results from the cartilage thickness mapping module. (d) Cartilage subregions from the cartilage parcellation module.
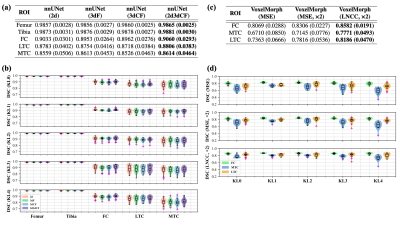
Figure 3. Performance of segmentation network and registration network. (a & b) The performance of nnU-Net variants evaluated by the Dice similarity coefficient (DSC). (c & d) The performance of template-to-image registration evaluated by the DSC between the template segmentation mask and individual segmentation mask.
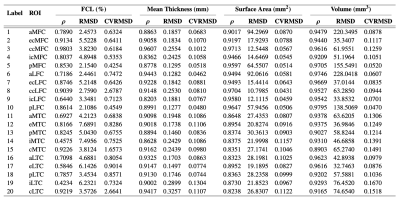
Figure 4. The effectiveness of the segmentation model evaluated by the correlation between regional metrics from manual labels and those from model segmentation. The Pearson’s correlation coefficient $$$\rho$$$, the root mean squared deviation (RMSD), and the coefficient of variation of RMSD (CVRMSD) are evaluated for each of the full-thickness cartilage loss (FCL), mean thickness, surface area, and volume.
DOI: https://doi.org/10.58530/2023/2457