2444
Deep learning–accelerated HASTE for single breath-hold liver T2-weighted imaging1Radiology, Zhongshan Hospital, Fudan University, Shanghai, China, 2MR Collaboration, Siemens (Shenzhen) Magnetic Resonance Ltd., Shenzhen, China, 3MR Application Predevelopment, Siemens Healthcare GmbH, Erlangen, Germany, Erlangen, Germany
Synopsis
Keywords: Liver, Data Acquisition, Deep-learning reconstruction
The HASTE sequence accelerated by deep learning (DL) reconstruction was used to perform liver T2-weighted imaging under a single breath-hold in a daily routine. Its image quality, including signal-to-noise ratio, contrast-to-noise ratio, artifacts, edge sharpness and slice continuity, was evaluated by comparing it with the conventional multi-breath-hold BLADE sequence. The result demonstrated that DL-accelerated HASTE could shorten the acquisition time remarkably while maintaining clinically satisfactory image quality.Introduction
T2-weighted imaging (T2WI) is one of the essential modalities in liver magnetic resonance imaging (MRI). Conventional T2WI is usually performed with respiratory triggering or using multiple breath-holds. Thus, the scan time is long, and inconsistency of the respiratory movement pattern also affects the image quality [1]. Applying the BLADE sequence can reduce breathing artifacts [2], but respiratory triggering or multi-breath-hold combinations are still needed for complete whole liver imaging. Also, the inconsistency of liver position in each breath-hold affects the continuity of image acquisition, thus affecting the overall image quality and the observation of lesions. Besides compressed sensing and parallel imaging, deep learning (DL) reconstruction has been developed recently to further increase the imaging speed, which has been demonstrated as clinically feasible [3-6]. This study applied DL reconstruction to accelerate the Half-Fourier Acquisition Single-shot Turbo spin Echo imaging (HASTE) sequence and thus implement single breath-hold liver T2W imaging. The clinical feasibility of single breath-hold DL-accelerated HASTE (DL-HASTE) in liver T2W imaging was verified by comparing it with the conventional multi-breath-hold BLADE sequence.Methods
This study enrolled 20 consecutive patients (14 male and 6 female) with liver space-occupying lesions (average age 57.3 ± 12.8 years). Patients with metal implants, hepatic cystic lesions, and contrast allergies were excluded. MRI examinations were performed on a 3T MR scanner (MAGNETOM Prisma; Siemens Healthcare, Erlangen, Germany) with an 18-channel body coil and a 32-channel integrated spine coil as receivers. The MR protocols included the research application T2W DL-HASTE, T2W BLADE, diffusion-weighted imaging (DWI), and pre- and post-contrast T1-weighted imaging (T1WI). Image reconstruction for DL-HASTE comprised a fixed iterative reconstruction scheme or variational network [7, 8], alternating between data consistency and a Convolutional Neural Network (CNN)-based regularization [3]. And the network was well trained using conventional HASTE images from volunteer acquisitions on various clinical 1.5T and 3T scanners (MAGNETOM scanners, Siemens Healthcare, Erlangen, Germany) [3]. Table 1 presents the parameters of DL-HASTE and BLADE sequences. All images were analyzed at the syngo.via workstation (Siemens Healthcare, Erlangen, Germany). The image quality, including artifacts and edge sharpness, was rated with a 5-point Likert-type scale (1 = very poor; 2 = poor; 3 = moderate; 4 = good; and 5 = excellent). Slice continuity was rated with a 2-point Likert-type scale (1 = slice discontinuity; 0 = no discontinuity). Signal-to-noise ratio (SNR), contrast-to-noise ratio (CNR), and long diameters of the lesion were measured for both sequences. The scores of image qualities were compared using the Wilcoxon rank-sum test. Long diameter, SNR and CNR were compared using the t test. P < 0.05 was considered statistically significant. Additionally, the agreement between the two sequences on the long-diameter measurement was evaluated by using Bland-Altman analysis.Results
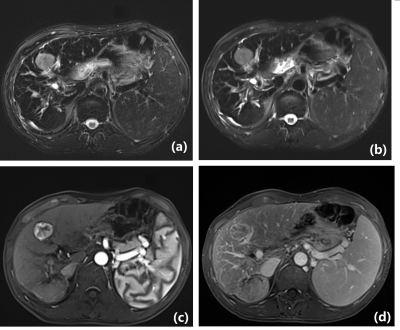
A total of 26 lesions were detected with DL-HASTE and 25 lesions with BLADE in 20 patients. The one lesion displayed in DL-HASTE but not in BLADE images was detected in both DWI and contrast-enhanced T1WI. The SNR and CNR of lesions were considerably higher in the DL-HASTE group than in the BLADE group (SNR: 172.85 ± 58.89 vs 44.75 ± 19.56, P < 0.001; CNR: 328.57 ± 273.19 vs 125.59 ± 101.83, P < 0.001) (figure 1). The artifact scores were higher in the DL-HASTE group than in the BLADE group. The scores of the edge sharpness were substantially lower in the DL-HASTE group than in the BLADE group (P < 0.05) (Table 2), but both of them were clinically acceptable. Slice discontinuity was detected in 2 of 20 patients for DL-HASTE and in 10 of 20 patients for BLADE. In the example case displayed in figure 2, the lesion indicated by the white arrow was hardly seen in the BLADE image, whereas it appeared clearly in both DL-HASTE and contrast-enhanced T1WI. No significant difference was observed in the measured diameters of the lesions between the 2 groups (P = 0.4). Bland-Altman analysis showed the average difference in the measured diameters was 0.52%, and the 95 % limits of agreement were 8.43%/-7.40% (figure 3). One representative example is shown in figure 4.Discussion and Conclusion
We used DL-HASTE to implement single breath-hold liver T2WI. The qualitative and quantitative evaluation of the acquired images demonstrated that the image quality achieved using DL-HASTE was better than that obtained using the multi-breath-hold BLADE sequence, except for the edge sharpness. However, the lower score of the edge sharpness did not affect the lesion detection capabilities for DL-HASTE. On the contrary, better slice continuity of DL-HASTE contributed by the single breath-hold improved the lesion detection capability, which was demonstrated by the example case shown in figure 1 and the result that DL-HASTE could detect more lesions than BLADE. In conclusion, DL-HASTE shortened the acquisition time of liver T2WI while maintaining clinically satisfactory image quality and lesion detection capability. Hence, DL-HASTE has the potential for use in clinical practice.Acknowledgements
No acknowledgement found.References
[1] Schreiber-Zinaman J, Rosenkrantz AB. Frequency and reasons for extra sequences in clinical abdominal MRI examinations. Abdom Radiol (NY) 2017;42:306–11. https://doi.org/10.1007/s00261-016-0877-6.
[2] Rosenkrantz AB, Mannelli L, Mossa D, Babb JS. Breath-hold T2-weighted MRI of the liver at 3 T using the BLADE technique: impact upon image quality and lesion detection. Clin Radiol 2011;66:426–33. https://doi.org/10.1016/j.
[3] Herrmann, J., Lingg, A., Kündel, M., Gassenmaier, S., Afat, S., Al-Mansour, H., Nickel, D., Koerzdoerfer, G., & Othman, A.E. (2021). Clinical Implementation of Deep Learning-Accelerated HASTE and TSE.
[4] Herrmann, J., Nickel, D., Mugler, J.P., Arberet, S., Gassenmaier, S., Afat, S., Nikolaou, K., & Othman, A.E. Development and Evaluation of Deep Learning-Accelerated Single-Breath-Hold Abdominal HASTE at 3 T Using Variable Refocusing Flip Angles. Investigative Radiology, 2021,56, 645 - 652.
[5] Herrmann J, Gassenmaier S, Nickel D, Arberet S, Afat S, Lingg A, Kündel M, Othman AE. Diagnostic Confidence and Feasibility of a Deep Learning Accelerated HASTE Sequence of the Abdomen in a Single Breath-Hold. Investigative Radiology. 2021 May 1;56(5):313-319.
[6] Sheng RF, Zheng LY, Jin KP, et al. Single-breath-hold T2WI liver MRI with deep learning-based reconstruction: A clinical feasibility study in comparison to conventional multi-breath-hold T2WI liver MRI. Magn Reson Imaging. 2021;81:75-81. doi:10.1016/j.mri.2021.06.014
[7] Hammernik K, Klatzer T, Kobler E, Recht MP, Sodickson DK, Pock T, et al. Learning a variational network for reconstruction of accelerated MRI data. Magn Reson Med. 2018 Jun;79(6):3055–3071.
[8] Schlemper J, Caballero J, Hajnal JV, Price AN, Rueckert D. A Deep Cascade of Convolutional Neural Networks for Dynamic MR Image Reconstruction. IEEE Trans Med Imaging. 2018 Feb;37(2):491–503
Figures