2424
Evaluation of renal function in chronic kidney disease using histogram analysis based on diffusion multi-models1Department of Radiology, Guangzhou Panyu Center Hospital, Guangzhou, China, 2Siemens Healthineers Ltd, Guangzhou, China
Synopsis
Keywords: Radiomics, Diffusion/other diffusion imaging techniques, Histogram analysis;magnetic resonance imaging;Renal function
This study established and validated a predictive model based on histogram features of four diffusion models to identify and evaluate early renal impairment in CDK. The results suggested that the model based on the ADC and MK could distinguish the normal and mild CDK well, and could accurately and noninvasively evaluate and predict early CDK renal dysfunction.Introduction/ purpose
It had always been an urgent clinical need to find an accurate and non-invasive method to evaluate the renal damage of chronic kidney disease (CDK). In recent years, more and more studies had been conducted on functional magnetic resonance imaging (MRI) in CDK. Monoexponential, biexponential (intravoxel incoherent motion, IVIM), stretched-exponential (SEM), and kurtosis (DKI), as advanced diffusion models, had been gradually applied to the study of kidney disease, and the results showed that the measures of these models could accurately evaluate the renal function [1-4]. However, previous studies on diffusion models focused on the average values of region of interest (ROI), which was limited in the evaluation of renal function. Histogram analysis using mathematical methods to analyze the distribution of voxel intensities within an image ROI could further provide more potential information and reflect the histological characteristics and heterogeneity [5]. In recent years, histogram analysis had made great progress in evaluating lesion and organ heterogeneity [6-10]. At present, there are relatively few studies on histogram analysis of CKD. The purpose of this study was to explore the radiomics features highly correlated with early kidney injury in CKD based on the histogram analysis of diffusion multi-models, and establish the early assessment and prediction model of CKD renal function.Method
This study included 49 patients with CKD (mild group, eGFR≥ 60 ml/min/1.73m2) and 25 healthy controls (HCs). All patients underwent diffusion weighted imaging (DWI) on a 3T MR scanner (MAGNETOM Prisma, Siemens Healthcare, Erlangen, Germany).The scanning parameters were as follows: TR=1500ms, TE= 66ms, FOV= 380 x 380 mm2, Voxel size=1.4 x 1.4 x 4.0 mm3, matrix=134 x 134, b values = 0,20, 50, 80, 150, 300, 500, 800, 1000, 1500, 2000, and 2500s/mm2. The measures of monoexponential (apparent diffusion coefficient [ADC]), IVIM (fast diffusion coefficient [Df], slow diffusion coefficient [Ds], and fraction of fast diffusion [f]), SEM (distributed diffusion coefficient [DDC] and anomalous exponent term [α]), and DKI (mean diffusivity [MD] and mean kurtosis [MK]) were calculated by an in-house developed software (BoDiLab) based on Python 3.7. ROI was manually drawn on the right proximal hilar of b0 image, which included the whole renal parenchyma, and avoided renal sinus tissue and artifacts. A total of 198 histogram features based on these diffusion measures of each ROI including shape-based and first order statistic features were extracted by OCIA software developed in-house. To early identify and predict renal function damage, 51 cases was selected as the training data set (34 patients, 17 HCs ) to learn and establish machine learning models,and another 23 cases as the independent testing data set (15 patients, 8 HCs) to evaluate the performance of the predictive models. Firstly, in order to balance data set of patients and HCs, up-samples of data set were performed by repeating random cases, and the normalization on the feature matrix were performed, since the numerical values calculated by different features are quite different. Then the dimension of the feature space was reduced by pearson correlation coefficients (PCC) methods and each feature was independent to each other. Before build the model, we used Kruskal Wallis to select significant features corresponding to the renal function damage. Logistic regression was used as the classifier that combines all the features. To determine the hyper-parameter (e.g. the number of features) of model, we applied cross validation with 5-fold on the training data set. The performance of the model was evaluated using receiver operating characteristic (ROC) curve analysis. The accuracy, sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of the models were also calculated at a cutoff value that maximized the value of the Yorden index. All above processes were implemented with FeAture Explorer Pro (FAE, V 0.5.4) on Python (3.7.6)Result
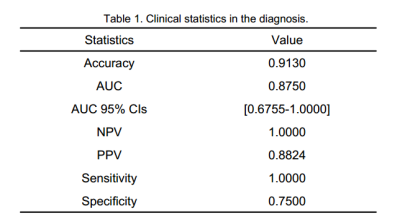
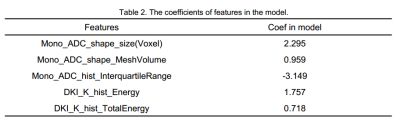
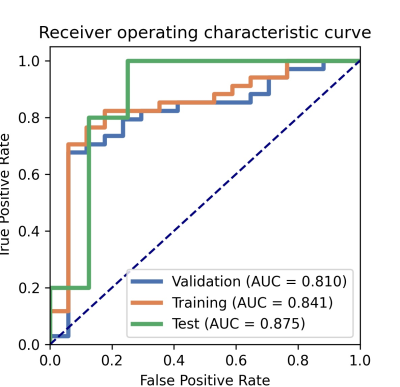
The model based on 5 features could get the highest AUC to classify participants between mild group and control group, with an AUC of 0.88, sensitivity and specificity of 94% and 75%, respectively. The clinical statistics in the diagnosis and the selected features were shown in Table 1 and Table 2. The ROC curve was shown in Figure 1.Discussion
This was a prospective study of histogram analysis based on four diffusion models to predict and evaluate renal function in CDK. The results showed that the predictive model based on the histogram features of ADC and MK was effective in differentiating the normal group from the mild group. Monoexponential model and DKI were effective and potential methods for non-invasive assessment of kidney damage in CKD, and their derived histogram parameters could be used as biological markers for potential assessment of kidney damage in CKD. However, the sample size of this study was small and no external validation was conducted, which should be further improved in future studies.Conclusion
The predictive model based on histogram features of diffusion multi-models could well distinguish and predict renal impairment in CKD, which was an effective and potential means of non-invasive assessment of renal impairment in CKD.Acknowledgements
No acknowledgement found.References
[1] Xu X, Palmer SL, Lin X, et al. Diffusion-weighted imaging and pathology of chronic kidney disease: initial study. Abdom Radiol (NY). 2018, 43(7):1749-1755. doi: 10.1007/s00261-017-1362-6.
[2] Mao W, Zhou J, Zeng M, et al. Chronic kidney disease: Pathological and functional evaluation with intravoxel incoherent motion diffusion-weighted imaging. J Magn Reson Imaging. 2018,47(5):1251-1259. doi: 10.1002/jmri.25861.
[3] Zhang J, Suo S, Liu G, et al. Comparison of Monoexponential, Biexponential, Stretched-Exponential, and Kurtosis Models of Diffusion-Weighted Imaging in Differentiation of Renal Solid Masses. Korean J Radiol. 2019,20(5):791-800. doi: 10.3348/kjr.2018.0474.
[4] Liu Y, Zhang GM, Peng X, et al. Diffusion kurtosis imaging as an imaging biomarker for predicting prognosis in chronic kidney disease patients. Nephrol Dial Transplant.2022,37(8):1451-1460. doi: 10.1093/ndt/gfab229.
[5] Lubner MG, Smith AD, Sandrasegaran K, et al. CT Texture Analysis: Definitions, Applications, Biologic Correlates, and Challenges. Radiographics. 2017,37(5):1483-1503. doi: 10.1148/rg.2017170056.
[6] Ding J, Xing Z, Jiang Z, et al. Evaluation of renal dysfunction using texture analysis based on DWI, BOLD, and susceptibility-weighted imaging. Eur Radiol. 2019,29(5):2293–2301. doi: org/ 10. 1007/ s00330- 018- 5911-3.
[7] Li A, Xing W, Li H, et al. Subtype Differentiation of Small (≤ 4 cm) Solid Renal Mass Using Volumetric Histogram Analysis of DWI at 3-T MRI. AJR Am J Roentgenol. 2018,211(3):614-623. doi: 10.2214/AJR.17.19278.
[8] Choi IY, Yeom SK, Cha J, et al. Feasibility of using computed tomography texture analysis parameters as imaging biomarkers for predicting risk grade of gastrointestinal stromal tumors: comparison with visual inspection. Abdom Radiol (NY) 2019,44(7):2346-2356. doi: 10.1007/s00261-019-01995-4.
[9] Ai Z, Han Q, Huang Z, et al. The value of multiparametric histogram features based on intravoxel incoherent motion diffusion-weighted imaging (IVIM-DWI) for the differential diagnosis of liver lesions. Ann Transl Med. 2020 Sep;8(18):1128. doi: 10.21037/atm-20-5109.
[10] Gao A, Zhang H, Yan X, et al. Whole-Tumor Histogram Analysis of Multiple Diffusion Metrics for Glioma Genotyping. Radiology. 2022 ,302(3):652-661. doi: 10.1148/radiol.210820.