2408
Continuous automated MRS data analysis workflow
Aaron T. Gudmundson1,2, Helge J. Zöllner1,2, Christopher W. Davies-Jenkins1,2, Erik G. Lee3,4, Timothy J. Hendrickson3,4, Richard A. E. Edden1,2, and Georg Oeltzschner1,2
1The Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2Kennedy Krieger Institute, F. M. Kirby Research Center for Functional Brain Imaging, Baltimore, MD, United States, 3Masonic Institute for the Developing Brain, University of Minnesota, Minneapolis, MN, United States, 4Informatics Institute, University of Minnesota, Minneapolis, MN, United States
1The Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2Kennedy Krieger Institute, F. M. Kirby Research Center for Functional Brain Imaging, Baltimore, MD, United States, 3Masonic Institute for the Developing Brain, University of Minnesota, Minneapolis, MN, United States, 4Informatics Institute, University of Minnesota, Minneapolis, MN, United States
Synopsis
Keywords: Software Tools, Spectroscopy, Automation
Large-scale application of magnetic resonance spectroscopy (MRS) is limited by demanding data processing and reliance on expert knowledge. We have developed an open-source continuous automated analysis workflow for MRS data, substantially reducing the effort for manual data analysis and quality control. This workflow is of particular interest for application-oriented non-expert users and large-scale multi-center multi-vendor MRS studies, and should substantially protect against loss of data from acquisition protocol errors.Introduction
Magnetic resonance spectroscopy (MRS) measures levels of endogenous biochemicals in-vivo with a clinical MRI. MRS analysis workflows are commonly not automated and require local spectroscopy expertise to be executed1. Applied research studies with MRS in the acquisition protocol require manual data organizing, amendment of analysis scripts, execution of the analysis pipeline, and assessment of processing success on individual datasets. Manual analysis labor is therefore a strongly rate-limiting factor that prevents the deployment of MRS at scale.The only reliable way to run MRS studies is to manually analyze each dataset after it has been acquired. Very few have the bandwidth and discipline to implement this workflow, and as a result, any change in acquisition protocol (or radiographer implementation of the protocol) goes unnoticed, too often resulting in loss of data. Here, we describe a fully automated data review and end-to-end analysis workflow to support non-expert users of MRS conducting local research studies.
Methods
The backbone of the workflow (Figure 1) is the main.py Python script executing raw data conversion, data analysis, and reporting.Raw data conversion using NIfTI-MRS & BIDS-MRS
NIfTI-MRS was developed to overcome longstanding issues with different proprietary raw data formats2. It is based on the NIfTI-2 standard, widely used in neuroimaging. Raw data is stored in well-defined array dimensions, accompanied by standardized header fields. The Brain Imaging Data Structure (BIDS) is a neuroimaging data organization standard3. File and folder naming conventions create a standardized hierarchy (study, subject, session, modality) that supports the automation of analysis pipelines, and data sharing. BIDS further catalogues necessary metadata on demographics and technical details. BIDS-MRS is an MRS-specific extension of BIDS currently in development4.
In our workflow, data conversion is performed with BIDScoin5, an automated tool that sorts source data according to DICOM metadata and file paths to determine ‘maps’, i.e., well-defined procedures for BIDS-compliant file naming and metadata annotation. These translation heuristics were created for one subject (sub-001) and summarized as YAML-formatted bidsmap. The bidsmap is then used thereafter as a template to populate newly acquired subject folders (sub-001, sub-002, etc). After sorting, the data is automatically converted into NIfTI format using dcm2niix for images and spec2nii for MRS data.
Data analysis in Osprey
Osprey is a MATLAB-based modular open-source software performing consensus-recommended modern MRS analysis6. For this work, we compiled Osprey v.2.4.0 as standalone executables for Windows 10 and MacOS using the MATLAB Compiler, including fully-automatic installation routines. Osprey requires a JSON-formatted7 job file specifying the analysis options, settings, and path definitions to the NIfTI-MRS/BIDS formatted data. This job file is automatically generated by the main.py script following a predefined template including settings and the e-mail addresses for reporting, which subsequently executes the analysis.
Reporting
Once Osprey has completed the requested analysis, a standardized HTML-reporting page is packaged into a zip file and attached to an e-mail sent to a set of addresses specified by the user in the job file template. The report contains visualizations of the different analysis stages (raw data loading, pre-processing, modeling, co-registration, segmentation) and quality control metrics (linewidth, SNR, fit error, frequency drift).
Source data directory monitoring
The backbone script main.py can be triggered manually after a new dataset has been added to the source data folder. We further increased the degree of analysis automation with an open-source file-watching service (watchman) to continuously monitor the source data directory for changes, and trigger main.py if new source data is added.
Results
The presented workflow was successfully used to process a test dataset including seven subjects. After initial configuration of the bidsmap, Osprey job file templates, and watchman scripts, no intervention was required, and data were recognized and analyzed when they were added to the ‘watched’ directory. The final summary report from the email is shown in Figure 2. Other Osprey routine outputs are automatically stored in the BIDS-specified derivatives folder.Discussion
We have established an automated workflow for project folder organization, data analysis, and reporting for applied MRS studies. Automation of MRS data analysis can improve reproducibility and efficiency, particularly at sites that have not previously established local workflows. Standardized workflows and data storage conventions also increase reusability of existing analysis code.The workflow is inherently modular and alternative approaches can easily supplement different parts of the workflow. For example, existing local conversions scripts to generate BIDS structures can be included. The analysis software can be replaced with other BIDS-compliant solutions such as FSL-MRS8, spant9, Vespa10 and jMRUI11. LCModel12 does not support NIfTI-MRS but could be integrated by FID-A13 pre-processing or calling LCModel from within Osprey.
Quality control metrics remain a challenge for MRS. Model residuals, Cramer-Rao Lower Bounds, linewidth, and SNR all provide valuable information14,15, but often fail to reliably indicate the presence of major artefacts like lipid contamination or out-of-voxel echoes. Visual inspection of individual spectra, therefore, continues to have a role, until complete set of QC metrics is developed and validated.
Conclusion
We present an open-source end-to-end workflow for continuous automated analysis of MRS data, making use of recent advances in standardized file format and data storage conventions. Reduced manual analysis overhead can help simplify the integration of MRS into large-scale multi-modal imaging studies and clinical trials.Acknowledgements
This work has been supported by NIH grants R00 AG062230, R21 EB033516, R01 EB016089, R01 EB023963, and P41 EB031771.References
- Soher BJ, Clarke WT, Wilson M, Near J, Oeltzschner G. Community-Organized Resources for Reproducible MRS Data Analysis. Magn Reson Med. 2022;88(5):1959-1961. doi:10.1002/mrm.29387
- Clarke WT, Bell TK, Emir UE, et al. NIfTI-MRS: A standard data format for magnetic resonance spectroscopy. Magn Reson Med. 2022;88(6):2358-2370. doi:10.1002/mrm.294183. Gorgolewski KJ, Auer T, Calhoun VD, et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci Data. 2016;3(1):160044. doi:10.1038/sdata.2016.44
- BIDS Extension Proposal 22 (BEP022): MRS (Magnetic Resonance Spectroscopy). Google Docs. Accessed November 4, 2022. https://docs.google.com/document/d/1pWCb02YNv5W-UZZja24fZrdXLm4X7knXMiZI7E2z7mY/edit?usp=sharing&usp=embed_facebook&usp=embed_facebook
- Zwiers MP, Moia S, Oostenveld R. BIDScoin: A User-Friendly Application to Convert Source Data to Brain Imaging Data Structure. Front Neuroinformatics. 2022;15. Accessed November 4, 2022. https://www.frontiersin.org/articles/10.3389/fninf.2021.770608
- Oeltzschner G, Zöllner HJ, Hui SCN, et al. Osprey: Open-source processing, reconstruction & estimation of magnetic resonance spectroscopy data. J Neurosci Methods. 2020;343:108827. doi:10.1016/j.jneumeth.2020.108827
- Pezoa F, Reutter JL, Suarez F, Ugarte M, Vrgoč D. Foundations of JSON schema. In: Proceedings of the 25th International Conference on World Wide Web. ; 2016.
- Clarke WT, Stagg CJ, Jbabdi S. FSL-MRS: An end-to-end spectroscopy analysis package. Magn Reson Med. 2021;85(6):2950-2964. doi:10.1002/mrm.28630
- Wilson M. spant: An R package for magnetic resonance spectroscopy analysis. J Open Source Softw. 2021;6(67):3646. doi:10.21105/joss.03646
- Soher BJ, Semanchuk P, Todd D, Steinberg J, Young K. VeSPA: Integrated applications for RF pulse design, spectral simulation and MRS data analysis. In: 19th Annual Meeting of the International Society for Magnetic Resonance in Medicine (ISMRM). ; 2011. Accessed May 19, 2020. https://cds.ismrm.org/protected/11MProceedings/files/1410.pdf11. Stefan D, Cesare FD, Andrasescu A, et al. Quantitation of magnetic resonance spectroscopy signals: the jMRUI software package. Meas Sci Technol. 2009;20(10):104035. doi:10.1088/0957-0233/20/10/104035
- Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30(6):672-679. doi:10.1002/mrm.1910300604
- Simpson R, Devenyi GA, Jezzard P, Hennessy TJ, Near J. Advanced processing and simulation of MRS data using the FID appliance (FID-A)—An open source, MATLAB-based toolkit. Magn Reson Med. 2015;77(1):23-33. doi:10.1002/mrm.2609114. Pedrosa de Barros N, Slotboom J. Quality management in in vivo proton MRS. Anal Biochem. 2017;529:98-116. doi:10.1016/j.ab.2017.01.017
- Tensaouti F, Desmoulin F, Gilhodes J, et al. Quality control of 3D MRSI data in glioblastoma: Can we do without the experts? Magn Reson Med. 2022;87(4):1688-1699. doi:10.1002/mrm.29098
Figures
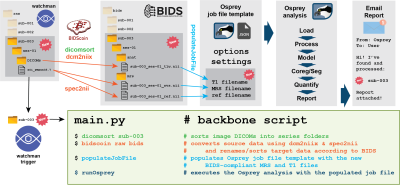
Illustration of the automated MRS data analysis workflow. The watchman service notices a new folder in the raw data directory that it monitors. It triggers the backbone script, which first uses dicomsort to sort image DICOM into series folders, then executes bidscoin which uses the dcm2niix and spec2nii programs to convert the raw data to NIfTI/NIfTI-MRS/BIDS format. The script then populates a template job file with paths to the new data and passes the job file on to an Osprey analysis instance. The last Osprey step sends a confirmation e-mail to specified addresses.
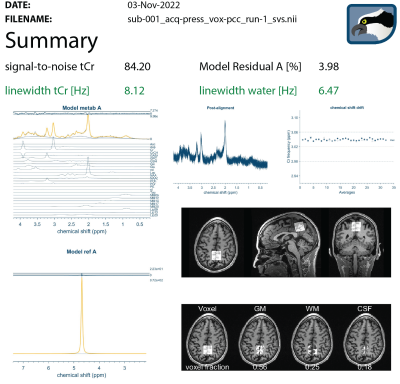
Summary dashboard reporting on all key steps of the automated MRS data analysis workflow (lightly edited from HTML output to minimize whitespace for presentation purposes). The HTML report is automatically e-mailed to a user-specified address after completion of the Osprey analysis. The summary includes quantitative quality control metrics (linewidth, SNR, model fit error) and visualizations of the processing, modeling, and co-registration/segmentation steps of the automated Osprey pipeline.
DOI: https://doi.org/10.58530/2023/2408