2406
QMRI-neuropipe: A flexible software framework for the analysis of quantitative MRI data1Pediatrics, University of Wisconsin–Madison, Madison, WI, United States, 2Medical Physics, University of Wisconsin–Madison, Madison, WI, United States, 3Waisman Center, University of Wisconsin–Madison, Madison, WI, United States
Synopsis
Keywords: Software Tools, Data Processing
Quantitative MRI provides a unique opportunity to characterize the underlying tissue and establish measurements that canserve as biomarkers. However, many of these methods require specialized workflows and and tools which limit their broader adoption. Here, we present QMRI-neuropipe, an open-source, flexible framework that provides a wide selection of methods, algorithms, and tools for processing and analyzing multiple qMRI datatypes. QMRI-neuropipe supports the Brain Imaging Datset Standard (BIDS) and currently supports processing of diffusion and relaxometry datasets. Future developments will aim to incorporate alternative quantitative MRI methods (e.g. MT, QSM, etc.) into the QMRI-neuropipe framework.Introduction
Quantitative magnetic resonance imaging (qMRI) aims to measure meaningful parameters that can be used to characterize the underlying tissue microstructure. The quantitative nature of these data suggest that derived measures are ideally reproducible within and across different sites, allowing qMRI data to be readily compared across different anatomical regions, among different subjects, and across time. As such, there is growing interest and use of qMRI techniques for a broad range of clinical and research applications. However, the processing of qMRI data, including diffusion MRI (dMRI) and relaxometry based techniques, among others, involves a number of steps prior to the estimation of the parametric maps; typically performed using tools developed in-house. This limits the standardization, reproducibility, and adoption of these techniques. Recent efforts to provide open-source software tools are underway (e.g. MRtrix1; qMRLab2; QUIT3, h-MRI4) and have allowed qMRI to be more accessible to the broader reseach and clinical community. Still, methods available within many of these toolboxes remain limited, making it difficult to capitalize on other available tools and therefore necessary to utilize multiple frameworks at a time. In this work, we build upon previous work and contribute an open-source, flexible framework, named QMRI-neuropipe, that provides a wide selection of methods and tools for processing and analyzing multiple qMRI datatypes. Specifically, we describe the current design and features of QMRI-neuropipe, provide representative use-cases of qmri-neuropipe, and discuss the potential of future developments.Design
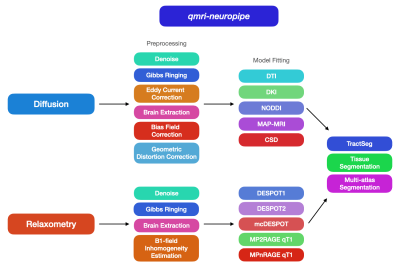
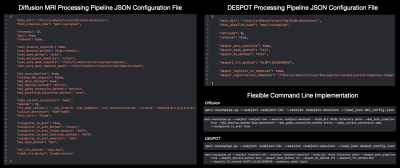
QMRI-neuropipe is a framework written in Python and is compatible across major operating systems, including macOS, Windows, and Linux. The choice of Python was made to allow easier implementation, sharing, and availability to the research community. An up-to-date version can be obtained via a GitHub repository (https://github.com/Developing-Brain-Imaging-Lab/qmri-neuropipe.git/). The use of Anaconda streamlines installation of required python dependencies and packages.QMRI-neuropipe includes a modular framework that allows existing methods to be flexibly combined into workflows and piplines, while also allowing new functionality to be easily integrated into the existing framework. QMRI-neuropipe integrates many tools and analysis routines from open-source neuroimaging software packages, including FSL5, AFNI6,7, MRtrix1, DIPY8, DMIPY9, the ANTsX ecosystem10, TractSeg11, among others. Moreover, the QMRI-neuropipe syntax allows a user to easily incorporate and interchange routines betweeen these other neuroimaging computing platforms. Where possible, QMRI-neuropipe supports multi-threaded and GPU based applications for faster processing, while integrations with the HTCondor software suite allows for the batch processing of a large number of jobs at one time.
Features
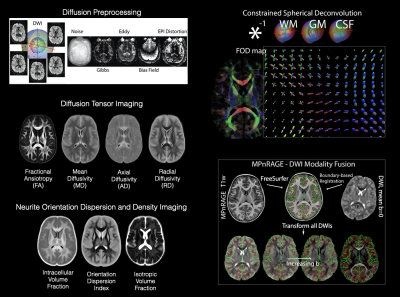
QMRI-neuropipe currently includes a collection of methods and algorithms for processing structural (T1w and T2w) MRI, diffusion MRI, and relaxometry data, as well as a variety of general image processing methods, including dicom-to-nifti conversion, brain masking, bias field correction, image smoothing, and image resampling. There is also an image normalization framework for performing linear and nonlinear registrations via FSL5 and ANTs10. qmri-neuropipesupports the brain imaging data structure (e.g. BIDS12), allowing pipelines to automatically configured based upon the data provided as well as providing a standardized organization of its subsequent derivatives and output files. Pipelines can also be configured using the JSON format, giving a user the flexibility to determine the methods and algorithms used by a given pipeline.Processing and Analyzing Diffusion MRI data. Pre-processing of dMRI data, including image denoising, Gibbs ringing artifact correction, eddy current and motion correction, and bias correction, are archieved utilizing wrappers around common methods from software packages of FSL, MRtrix, and DIPY. Additional tools for combining multi-phase encode DWI data and ensuring the number of DWIs agrees with the number of b-values and b-vectors ensure data consistency. qmri-neuropipe also includes several methods for geometric distortion correction, including FSL’s TOPUP13, B0 fieldmap based correction (e.g. FSL fugue5), and registration correction with an accompanying structural MRI. A number of diffusion models – DTI, DKI, NODDI, CSD, MAP-MRI – and optimization routines (e.g. ordinary least squares, weighted least squares, and nonlinear least squares) are provided using wrappers around models provided within MRtrix, DIPY, and DMIPY, thus giving a user the ability to adopt advanced dMRI models more easily. QMRI-neuropipe also includes additional tools for performing tract segmentation, diffusion tractography, bundle selection, and along tract segmentation.
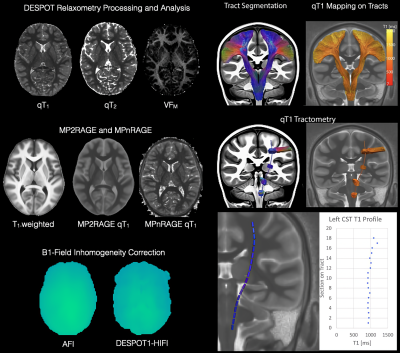
Processing and Analyzing Quantitative Relaxometry QMRI-neuropipe also supports the processing of several quantitative relaxometry models, including DESPOT1/DESPOT2/mcDESPOT techniques14,15, MP2RAGE16, and MPnRAGE17,18. DESPOT routines include the rigid registration of variable flip angle SPGR and bSSFP data and estimation of qT1, qT2 and myelin water fraction maps. Actual Flip-angle Imaging (AFI19) and DESPOT1-HIFI14 provide methods for correcting for B1 field inhomogeneity. Methods for normalization of relaxometry data as well as multi-modal data analysis (e.g. dMRI-Relaxometry) are additionally provided.
Conclusions
By combining native methods and cutting-edge techniques across available neuroimaging software packages, QMRI-neuropipe is able to provide a comprehensive, flexible framework for the processing and analysis of multiple types of qMRI data. Future developments will focus on the integration of alternative diffusion and relaxometry models as well as incorporating methods beyond diffusion and relaxometry. We envision QMRI-neuropipe serving as a powerful tool that enables qMRI to be more accessible to the neuroscience and clinical research communities and allows for further standardization, reproducibility, and adoption of these techniques.Acknowledgements
This work was supported by R00 MH11056 (Dr. Dean) from the National Institute of Mental Health, National Institutes of Health and R01NS123378 (Dr. Adluru) from the National Institute of Neurological Disease and Stroke, National Institutes of Health. Infrastructure support was also provided, in part, by grant U54 HD090256 from the Eunice Kennedy Shriver NICHD, National Institutes of Health (Waisman Center).References
1 Tournier, J. D. et al. MRtrix3: A fast, flexible and open software framework for medical image processing and visualisation. NeuroImage 202, 116137 (2019). https://doi.org:https://doi.org/10.1016/j.neuroimage.2019.116137
2 Karakuzu, A. et al. qMRLab: Quantitative MRI analysis, under one umbrella. Journal of Open Source Software 5, 2343 (2020).
3 C Wood, T. QUIT: QUantitative imaging tools. Journal of Open Source Software 3, 656 (2018).
4 Tabelow, K. et al. hMRI–A toolbox for quantitative MRI in neuroscience and clinical research. Neuroimage 194, 191-210 (2019).
5 Jenkinson, M., Beckmann, C. F., Behrens, T. E., Woolrich, M. W. & Smith, S. M. Fsl. Neuroimage 62, 782-790 (2012). https://doi.org:10.1016/j.neuroimage.2011.09.015
6 Cox, R. W. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Computers and Biomedical research 29, 162-173 (1996).
7 Cox, R. W. AFNI: what a long strange trip it's been. Neuroimage 62, 743-747 (2012). https://doi.org:10.1016/j.neuroimage.2011.08.056
8 Garyfallidis, E. et al. Dipy, a library for the analysis of diffusion MRI data. Front Neuroinform 8, 8 (2014). https://doi.org:10.3389/fninf.2014.00008
9 Fick, R. H. J., Wassermann, D. & Deriche, R. The Dmipy Toolbox: Diffusion MRI Multi-Compartment Modeling and Microstructure Recovery Made Easy. Frontiers in Neuroinformatics 13 (2019). https://doi.org:10.3389/fninf.2019.00064
10 Tustison, N. J. et al. The ANTsX ecosystem for quantitative biological and medical imaging. Scientific Reports
11, 1-13 (2021). 11 Wasserthal, J., Neher, P. & Maier-Hein, K. H. TractSeg-Fast and accurate white matter tract segmentation. NeuroImage 183, 239-253 (2018).
12 Gorgolewski, K. J. et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Scientific data 3, 1-9 (2016).
13 Andersson, J. L., Skare, S. & Ashburner, J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage 20, 870-888 (2003). https://doi.org:10.1016/S1053-8119(03)00336-7
14 Deoni, S. C. High-resolution T1 mapping of the brain at 3T with driven equilibrium single pulse observation of T1 with high-speed incorporation of RF field inhomogeneities (DESPOT1-HIFI). J Magn Reson Imaging 26, 1106-1111 (2007). https://doi.org:10.1002/jmri.21130
15 Deoni, S. C., Rutt, B. K., Arun, T., Pierpaoli, C. & Jones, D. K. Gleaning multicomponent T1 and T2 information from steady-state imaging data. Magn Reson Med 60, 1372-1387 (2008). https://doi.org:10.1002/mrm.21704
16 Marques, J. P. et al. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. NeuroImage 49, 1271-1281 (2010). https://doi.org:https://doi.org/10.1016/j.neuroimage.2009.10.002
17 Kecskemeti, S. & Alexander, A. L. Three-dimensional motion-corrected T1 relaxometry with MPnRAGE. Magnetic Resonance in Medicine 84, 2400-2411 (2020). https://doi.org:https://doi.org/10.1002/mrm.28283
18 Kecskemeti, S. et al. MPnRAGE: A technique to simultaneously acquire hundreds of differently contrasted MPRAGE images with applications to quantitative T1 mapping. Magn Reson Med 75, 1040-1053 (2016). https://doi.org:10.1002/mrm.25674
19 Yarnykh, V. L. Actual flip-angle imaging in the pulsed steady state: a method for rapid three-dimensional mapping of the transmitted radiofrequency field. Magn Reson Med 57, 192-200 (2007). https://doi.org:10.1002/mrm.21120
Figures