2404
PyGRASP: A standalone python image reconstruction library for DCE-MRI acquired with radial sampling1Quantitative Intelligent Imaging Lab (QUIN), Department of Radiology, Boston Children's Hospital and Harvard Medical School, Boston, MA, United States
Synopsis
Keywords: Software Tools, DSC & DCE Perfusion
Dynamic contrast enhanced MRI (DCE-MRI) is capable of quantitative assessment of renal function and evaluation of the detailed anatomy of the urinary tract and renal arteries in patients. To reconstruct the dynamic volumes in DCE-MRI, Golden-angle RAdial Sparse Parallel (GRASP) reconstruction algorithm is commonly used. In this software, we have developed an efficient open-source, purely Python-based, standalone GRASP reconstruction library called pyGRASP that allows researchers to access the source code for development, facilitating flexible deployment with readable code and no compilation, and easy utilization without requirement of a steep learning curve.Introduction
In dynamic contrast-enhanced (DCE) magnetic resonance imaging (MRI), a dynamic series of image stacks are acquired, capturing the passage of the contrast agent through the tissue compartments. Dynamic radial imaging with a golden angle stack of stars sampling trajectory has been increasingly used to acquire DCE-MRI data due to its advantages compared to dynamic cartesian imaging, such as being robust to breathing motion and achieving high temporal resolution imaging. To reconstruct a dynamic series of volumes at high temporal resolution (~3 seconds/volume), the data must be reconstructed from undersampled measurements. Golden-angle RAdial Sparse Parallel (GRASP) reconstruction algorithm1-3. uses temporal regularization to reduce undersampling artifacts in the form of streaking in radial imaging and can reconstruct a dynamic series of volumes at the desired temporal resolution. In this software, we have developed an efficient open-source, purely python-based, standalone GRASP reconstruction library called pyGRASP that allows researchers to access the source code for development, facilitating flexible deployment with readable code and no compilation, and easy utilization without requirement of a steep learning curve. This library includes preprocessing by extracting information from raw data acquired scanners and coil sensitivity map estimation, coil compression and coil removal options (to remove coils that generate artifacts)4 for improving speed and image quality, and image reconstruction using a compressed sensing approach based on GRASP technique, followed by post-processing. The proposed library has options to accelerate computations by parallel processing using either GPU or CPU resources. We also provide docker files for quick and easy deployment over any operating system or cloud. pyGRASP library is publicly available in GitHub5.Methods
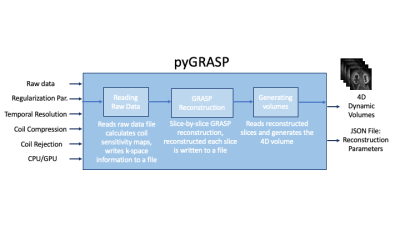
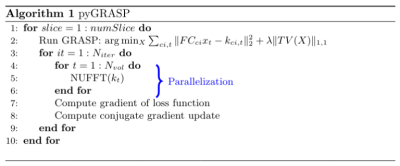
Methods: pyGRASP consists of three modules (Figure 1). The first reads the raw data acquired, writes the k-space data into a file, and calculates coil sensitivity maps based on the Walsh method6. The pyGRASP library provides options for coil rejection and coil compression that can be enabled or disabled by the user. For coil rejection, the quality of information each coil carries is evaluated based on a mutual information (MI) -based metric computed between a reference image (which is a combination of all data) and each coil image, and a set of coils, whose MI metric falls below the threshold are removed, reducing streaking artifacts and improving the SNR of the reconstructed image4. For coil compression, PCA is employed by compressing data from many channels into fewer virtual coils, with the goal of reducing computational time. In the second module, GRASP reconstruction is computed, solving $$$f(X) = ||FC x_t - k_t||_2^2 + \lambda ||TV(X)||_1^1$$$ where $$$X=\{x_t|t∈\{1,2,⋯,N\}\}$$$, F is the nonuniform Fourier transform (NUFT), C is the coil profile transform, TV(.) is the total variation, kt and xt are the k-space data, and image for a time frame t, respectively. The first term in the loss function is used for the data consistency between measurements and reconstructed data, and the second is for regularization in time. This second term minimizes the total variation of reconstructed volumes in the temporal direction used as a sparsifying transform, followed by the computation of its L1 norm for enforcing sparsity in time. Users can tune the weight of λ for the temporal regularization term (second term) and choose the number of spokes per volume to adjust the temporal resolution of reconstructed volumes. In pyGRASP, we implemented parallelization over dynamic volumes to reduce computation time as observed in the flowchart of the pyGRASP (Figure 2). We have measured computation time for MATLAB-based GRASP, CPU-based pyGRASP, and GPU-based pyGRASP for a dataset of 8 mins of data, including 2600 spokes, 21 slices, 38 channels, and a matrix size of 448. We have reconstructed dynamic images for different regularization parameters and temporal resolutions.Results
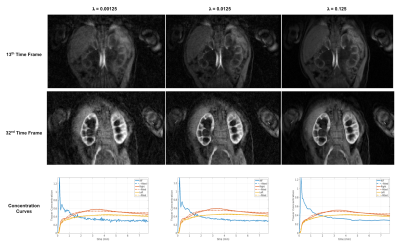
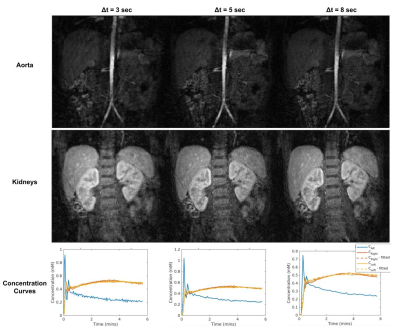
GPU parallelization reduced the computation time to ~3.5 mins/slice using a batch size of 10 (GPU’s memory was 24564MiB), whereas computation time was ~ 7.3 mins/slice when it was run on a CPU with 32 cores. On the other hand, the computation time of MATLAB-based GRASP was very long, with ~99 mins/slice for the same dataset. For one representative subject, reconstructed images for three different regularization parameters and extracted concentration curves are given in Figure 3. Results for three different temporal resolutions are given in Figure 4. The image quality improves when we increase the regularization coefficient. However, the temporal signal for the arterial input starts to smooth out for a large regularization coefficient7.Discussion and Conclusion
The pyGRASP library provides a python-based image reconstruction library specific for GRASP reconstruction in DCE-MR images that is easy to use. Source code, example raw data and readme files, and docker implementation for direct utilization are provided on the GitHub page5. Currently, the first raw data reading module is compatible only with data acquired in Siemens MRI scanners. As future work, we will enable the reading module’s compatibility with ISMRMRD format8.Acknowledgements
This work was supported in part by NIH grants R01 EB019483, R01 NS121657, R01 DK125561, R21 DK123569, R21 EB02962, S10OD025111 and a pilot grant (PP-1905-34002) from the National Multiple Sclerosis Society.References
1. Feng, Li, et al. "Golden angle radial sparse parallel MRI: combination of compressed sensing, parallel imaging, and golden-angle radial sampling for fast and flexible dynamic volumetric MRI." Magnetic resonance in medicine 72.3 (2014): 707-717.
2. Kurugol, S., Afacan, O., Lee, R.S. et al. Prospective pediatric study comparing glomerular filtration rate estimates based on motion-robust dynamic contrast-enhanced magnetic resonance imaging and serum creatinine (eGFR) to 99mTc DTPA. Pediatr Radiol 50, 698–705 (2020).
3. Coll Font, Jaume, et al. "Bulk motion-compensated DCE-MRI for functional imaging of kidneys in newborns." Journal of Magnetic Resonance Imaging 52.1 (2020): 207-216.
4. Kocanaogullari A, Ariyurek C, Afacan O, Kurugol S.Coil Selective Golden Angle DCE-MR Image Reconstruction using Mutual Dependence. In Proceedings of the 31st Joint Annual Meeting of ISMRM-ESMRMB, London, UK, 2022. p. 2442
5. Quin-med-harvard-edu. 2022. pyGRASP. https://github.com/quin-med-harvard-edu/pyGRASP
6. Inati, S. J., Hansen, M. S., & Kellman, P. (2014). A fast optimal method for coil sensitivity estimation and adaptive coil combination for complex images. Proceedings of the 22nd Annual Meeting of ISMRM, Milan, 4407.
7. S. Kurugol, O. Afacan, A. Stemmer, R. S. Lee, J. S. Chow, and S. K. Warfield, Glomerular filtration rate estimation by motion-robust high spatiotemporal resolution DCE-MRI with radial VIBE and comparison with plasma clearance of 99mTc-DTPA. Proc. of Int. Soc. of Magnetic Resonance Imaging, 2019.
8. Inati SJ, Naegele JD, Zwart NR, Roopchansingh V, Lizak MJ, Hansen DC, Liu CY, Atkinson D, Kellman P, Kozerke S, Xue H, Campbell-Washburn AE, Sørensen TS, Hansen MS. ISMRM Raw data format: A proposed standard for MRI raw datasets. Magn Reson Med. 2017 Jan;77(1):411-421.
Figures