2403
A diffusion-weighted MRI pulse sequence development toolbox in the open source GinkgoSequence framework1BAOBAB, NeuroSpin, Paris-Saclay University, CNRS, CEA, Gif-sur-Yvette, France, 2Mind, NeuroSpin, Paris-Saclay University, INRIA, CEA, Gif-sur-Yvette, France
Synopsis
Keywords: Software Tools, Pulse Sequence Design
Developing MRI pulse sequences demands to access proprietary sequences, to open up parts of the code provided by the manufacturer, and to deal with intellectual property issues. To address these limitations, we have provided the community with GinkgoSequence, a modular and open-source MRI pulse sequence development framework for Siemens devices. In this work, we present the addition of a new diffusion-weighted sequences development toolbox in the existing GinkgoSequence framework. It includes trapezoidal and free-waveform diffusion gradients, fat saturation, echo-planar reading, partial Fourier and GRAPPA accelerations, and stimulated echo acquisition mode (STEAM). To assess its quality, it has been tested in-vivo.Introduction
Diffusion-weighted MRI (dMRI) has become an established tool to map brain microstructure in vivo. Since the introduction of the Pulsed Gradient Spin Echo sequence1 (PGSE), various dMRI sequences have been proposed to offer innovative preparation, excitation, diffusion sensitization, or echo train read schemes. Unfortunately, MRI manufacturers generally propose a very limited set of diffusion sequences on their systems which satisfies most of the customers’ clinical needs. They sometimes give access to advanced diffusion sequences offering more complex gradient shapes, like OGSE2 and b-tensors3, or accelerated readout schemes shared by the community. However, it can be difficult to handle intellectual property issues or to get access to their codes to customize them further. To overcome these obstacles, among the existing open-source solutions for MRI pulse sequence development including the cross-platform Pulseq4 and the code-free SequenceTree5 frameworks which are not optimized for dMRI, we have recently developed GinkgoSequence6, an open-source solution for generic and object-oriented development of MRI pulse sequences. We demonstrate in this work how it can be exploited to propose a novel toolbox of dMRI pulse sequences with full access to their high quality and readability source codes.Material and methods
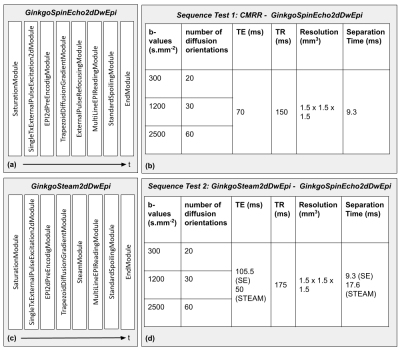
Modules development: In GinkgoSequence, we developed a panel of modules required to implement dMRI sequences, including trapezoidal and free-waveform diffusion gradients (like sine/cosine/trapezoid-sine/trapezoid-cosine, oscillating gradients2 or more general gradient waveforms3,7), saturation pulses, 2D and 3D EPI readouts including partial Fourier8 (PF) and GRAPPA9 accelerations, and a stimulated echo acquisition module (STEAM10). These tools allowed the quick development of a first set of standard dMRI pulse sequences such as 2D or 3D Diffusion-Weighted (DW) Spin-Echo EPI or 2D DW STEAM EPI to enable the exploration of longer diffusion times. The current class diagram of the GinkgoSequence framework is depicted in Fig.1, and its code is accessible online11. The source repository also includes example sequences, which were developed to be compatible with the Gadgetron12 toolkit for MRI data reconstruction.Sequence test 1 [GinkgoSpinEcho2dDwEpi - CMRR]: We benchmarked our GinkgoSequence toolbox with the dMRI sequence available from CMRR13, disabling its multi-band feature. This sequence implements a fat saturation pulse, a 2D excitation, an EPI reading, and PF/GRAPPA accelerations. Using the GinkgoSequence modules as described in Fig.2-a, we developed a similar GinkgoSpinEcho2dDwEpi sequence and performed an in vivo multiple-shell DW acquisition using the parameters described in Fig.2-b. This sequence was designed to loop over diffusion orientations provided in a text file along with the durations and timings for the DW gradients.
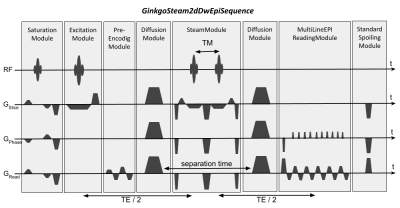
Sequence test 2 [GinkgoSteam2dDwEpi - GinkgoSpinEcho2dDwEpi]: The STEAM version of this sequence, GinkgoSteam2dDwEpi, was also implemented as described in Fig.2-c, and detailed in Fig.3. To assess its performance it was compared with the GinkgoSpinEcho2dDwEpi sequence. Given that the STEAM signal is halved compared to the equivalent spin-echo signal, the echo times were calculated to obtain an equivalent signal on the two sequences and minimized to obtain an acceptable acquisition time with the same DW parameters as test 1, leading to the table described in Fig.2-d.
Both tests were performed on a 3T Prisma Siemens system on a healthy volunteer and reconstructed using Gadgetron.
Post-processing: The obtained DW images were analyzed with the Ginkgo toolbox14 including the correction of imaging artifacts, the computation of a mask of the brain, the inference of diffusion maps from the diffusion tensor model15 and the analytical q-ball model16, and inference of the structural connectivity using a regularized deterministic tractography method17.
Quality Control: For both protocols, Fractional Anisotropy (FA) histograms, FA root-mean-square (RMS) error, and signal-to-noise (SNR) ratios computed on the b=0s.mm-2 reference scans were compared.
Results
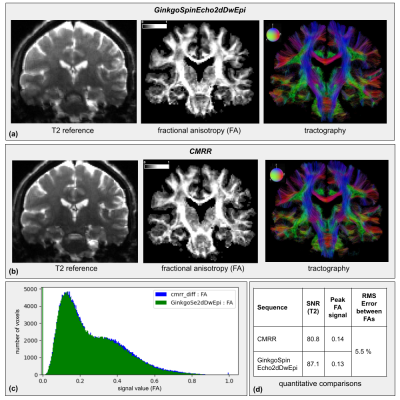
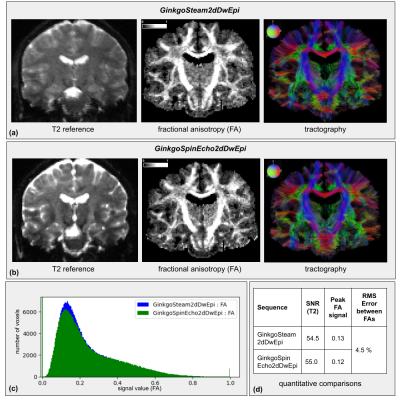
Results from sequence test 1 are presented in Fig.4-a&b, including a T2 reference, a FA map, and a tractography of the same slice for each sequence. FA histograms and a table providing the values of their peaks, as well as SNRs and RMS error, are displayed in Fig.4-c&d.The sequence test 2 results are shown similarly in Fig.5.
Discussion
The results from CMRR and GinkgoSpinEcho2d sequences were quite reproducible. The T2-weighted images showed an identical EPI deformation and T2 contrast. The FA maps were similar with a small RMS error, confirmed by the histogram, showing an equivalent distribution of its value over the image. The tractography also corroborated this result, highlighting the same trajectories for the corpus callosum and the cortico-spinal tracts. The obtained SNRs were almost equal, with the GinkgoSpinEcho2dDwEpi sequence showing a slightly higher SNR.The comparison of the STEAM and spin-echo sequences showed that the T2 images had different contrasts as expected from their different echo times, and presented the anticipated equivalent SNRs. Both FA maps and tractographies showed the detailed tracts of the brain, and the FA histograms also showed similar profiles with equivalent peaks and a small RMS error.
Conclusion
The GinkgoSequence framework allowed a modular, efficient, and open-source development of various dMRI pulse sequences which deliver top-quality dMRI data in vivo on human subjects. Future work will target the development of further modules to offer multiband features and parallel transmission techniques to meet the expectations of ultra-high-field imaging at 7T and 11.7T. It represents to our knowledge the first pulse sequences development framework enabling the efficient development of any dMRI pulse sequence.Acknowledgements
This work has been partially funded by the Phare Ph.D. Program of the CEA (France).
We would like to thank the RX operators from NeuroSpin’s UNIACT clinical team: Ginigsty C., Lecomte Y., Berland V., and Bériot L..
References
1. Stejskal E. O. and Tanner J. E.: Spin diffusion measurements: spin echoes in the presence of a time‐dependent field gradient. The journal of chemical physics, 1965; 42(1):288-292.
2. Van, A.T., Holdsworth, S.J., Bammer, R.: In vivo investigation of restricted diffusion in the human brain with optimized oscillating diffusion gradient encoding, Magnetic Resonance in Medicine, 2014; 71:83-94.
3. Topgaard, D.: Multidimensional diffusion MRI, Journal of Magnetic Resonance, 2017; 275:98-113.
4. Layton, K.J., et al.: Pulseq: a rapid and hardware‐independent pulse sequence prototyping framework, Magnetic Resonance in Medicine, 2017; 77(4):1544-1552.
5. Magland, J.F., Li, C., Langham, M.C. and Wehrli, F.W.: Pulse sequence programming in a dynamic visual environment: SequenceTree., 2016, Magnetic Resonance in Medicine; 75:257-265.
6. Artiges, A., et al.: Ginkgo: a novel modular and Open Source MRI pulse sequence development framework dedicated to MRI systems, proceedings of ISMRM 2022, 2022.
7. Drobnjak, I., Siow, B., Alexander, D.C.: Optimizing gradient waveforms for microstructure sensitivity in diffusion-weighted MR, Journal of Magnetic Resonance, 2010; 206(1):41-51.
8. Feinberg, DA., Hale, JD., Watts, JC., et al.: Halving MR imaging time by conjugation: demonstration at 3.5 kG., Radiology, 1986; 161:527-531.
9. Griswold, M.A., Jakob, P.M., Heidemann, R.M., et al.: Generalized autocalibrating partially parallel acquisitions (GRAPPA), Magnetic Resonance in Medicines, 2002; 47:1202-1210.
10. Kleinnijenhuis, M., Mollink, J., Lam, W.W., et al.: Choice of reference measurements affects quantification of long diffusion time behavior using stimulated echoes, Magnetic Resonance in Medicine, 2017; 79(2).
11. https://framagit.org/cpoupon/gkg/-/tree/master/pulse-sequence
12. Hansen, M. S., Sørensen, T. S.: Gadgetron: an open source framework for medical image reconstruction. Magnetic resonance in medicine, 2013; 69(6):1768-1776.
13. Setsompop, K., Cohen-Adad, J., Gagoski, B.A., Raij, T., Yendiki, A., Keil, B., Wedeen, V.J., Wald, L.L.: Improving diffusion MRI using simultaneous multi-slice echo planar imaging, Neuroimage, 2012; 63(1):569-80.
14. https://framagit.org/cpoupon/gkg
15. Basser, P.J., Mattiello, J., LeBihan, D.: MR diffusion tensor spectroscopy and imaging, Biophysical Journal, 1994; 66(1):259-267.
16. Descoteaux, M., Angelino, E., Fitzgibbons, S., Deriche, R.: Regularized, fast, and robust analytical Q-ball imaging, Magnetic Resonance in Medicine, 2007; 58(3):497-510.
17. Perrin, M., Poupon, C., Cointepas, Y., Rieul, B., Golestani, N., Pallier, C., Rivière, D., Constantinesco, A., Le Bihan, D., Mangin, JF.: Fiber tracking in q-ball fields using regularized particle trajectories, Inf Process Med Imaging., 2005; 19:52-63.
Figures