2398
CMRSeq - A Python package for intuitive sequence design1University and ETH Zurich, Zurich, Switzerland
Synopsis
Keywords: Software Tools, Pulse Sequence Design
We present CMRseq, a new python package for MR sequence definitions. The package builds upon successful concepts of previous frameworks, while leveraging Python functionality to increase usability and maintainability. CMRseq features physical unit checks, adheres to coding style conventions and includes unit-test coverage and continuous integration infrastructure for documentation. Import and export functionality to facilitate compatibility with popular formats is also included. The API reference as well as information on installation and how to get started is publicly available at: https://people.ee.ethz.ch/~jweine/cmrseq/latest/index.html
Keywords: Open source, reproducible science, sequence design, software tools, visualization
Introduction
Defining MR sequences without being bound to a vendor-specific framework plays a central role in simulation as well as in the efforts of promoting open and reproducible science. In recent years, multiple software packages have been proposed that aim at providing a vendor-agnostic framework for sequence definitions1-4. Recent work has made use of these frameworks to establish open source pipelines comparing measurements and simulations5-7. We feel that efforts to create community-driven open-source software projects are often hampered by missing infrastructure accompanying the actual implementation (e.g. accessible programming languages or documentation). To this end we have developed the software package CMRseq. While we built on some concepts used in the Pulseq/PyPulseq1,2 framework, our implementation makes more efficient use of Python language features to increase intuitiveness and usability. Additionally, emphasis was put on best practices concerning coding-style, separation of functionality and documentation.Design and Features
A schematic overview of the modules contained in the CMRseq package is provided in Fig 1.Sequence Definition
The core functionality is given by the following components:
- The ‘SystemSpec’ class: Enabling a consistent definition of system parameters and also providing system-specific functionality such as rounding times to a given raster or calculating the shortest possible gradient within system limits.
- The ‘Sequence’ class: Container for sequence building blocks (gradients, delays, RF and ADC-blocks).
- The ‘bausteine’-module: Providing definitions for the fundamental building blocks of MR sequences.
To compose a set of building blocks into a sequence, multiple methods are available. On instantiation of a sequence object the user can pass a list of building block instances, which can also be empty. Blocks as well as other sequence objects can be appended to an existing sequence object, which will automatically shift the added objects according to the end time of the sequence. The sequence class implements the addition operator, which means that sequence objects can be combined by using the “+” operation. All sequence operations can be performed in-place or by creating copies.
Sequence library
In addition to the core functionality, CMRseq includes a module that contains parametric definitions for recurring compositions of building blocks (i.e. slice selective gradients) as methods which return a 'Sequence' object. Due the mutable nature of the building blocks contained in a sequence, it is trivial to replace and/or modify elements in the 'Sequence' object. This allows straightforward iteration of sequence changes without the necessity of re-running the script for composing the sequence from building blocks. To structure the parametric definitions contained in the library, the methods are grouped into submodules corresponding to their use (e.g. excitation, readout or entire sequences).
Additional utility
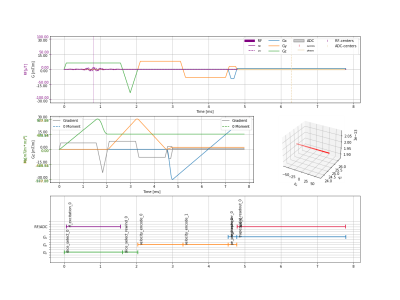
Plotting - The plotting module contains methods to illustrate sequence diagrams, k-space trajectories as well as the sequence structure showing the block names and timings. Similar to the popular plotting package seaborn9, it is possible to hand in existing axes in which the plots are inserted to conveniently create complex figures (Fig 2).
Exporting/Importing files - For saving and communicating the sequence definitions within the CMRseq framework an automatic export to JSON files is implemented. Furthermore, reading and writing the Pulseq file format is supported, to make use of the existing export to scanner functionality of Pulseq.
Example scenarios
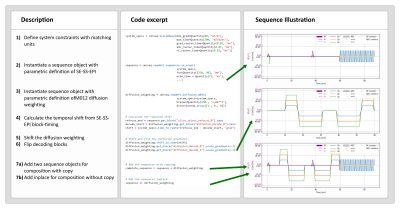
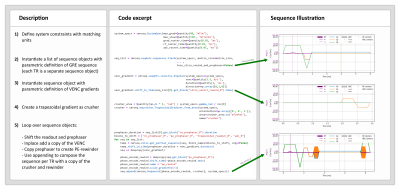
Figure 3 and 4 demonstrate two scenarios of defining a sequence. In the first scenario a SE-EPI is combined with diffusion weighting gradients, and in the second scenario velocity encoding gradients are added to a spoiled GRE.Conclusion
The proposed CMRseq package leverages and extends successful concepts of previous frameworks. Providing a clear software structure, established code style and continuous integration principles including automatic unit-testing, versioning and documentation builds, support the specific needs for fast sequence prototyping and open, reproducible science.Code: https://gitlab.ethz.ch/jweine/cmrseq
Documentation: https://people.ee.ethz.ch/~jweine/cmrseq/latest/index.html
Acknowledgements
References
- Layton KJ, Kroboth S, Jia F, et al. Pulseq: A rapid and hardware-independent pulse sequence prototyping framework. Magn. Reson. Med. 2017;77:1544–1552 doi: 10.1002/mrm.26235.
- Ravi K, Geethanath S, Vaughan J. PyPulseq: A Python Package for MRI Pulse Sequence Design. J. Open Source Softw. 2019;4:1725 doi: 10.21105/joss.01725.
- Nielsen JF, Noll DC. TOPPE: A framework for rapid prototyping of MR pulse sequences. Magn. Reson. Med. 2018;79:3128–3134 doi: 10.1002/mrm.26990.
- Cordes C, Konstandin S, Porter D, Günther M. Portable and platform-independent MR pulse sequence programs. Magn. Reson. Med. 2020;83:1277–1290 doi: 10.1002/mrm.28020.
- Loktyushin A, Herz K, Dang N, et al. MRzero - Automated discovery of MRI sequences using supervised learning. Magn. Reson. Med. 2021;86:709–724 doi: 10.1002/MRM.28727.
- Xanthis CG, Aletras AH. CoreMRI: A high-performance, publicly available MR simulation platform on the cloud. PLoS One 2019;14:1–26 doi: 10.1371/journal.pone.0216594.
- Wang G, Luo T, Nielsen JF, Noll DC, Fessler JA. B-Spline Parameterized Joint Optimization of Reconstruction and K-Space Trajectories (BJORK) for Accelerated 2D MRI. IEEE Trans. Med. Imaging 2022;41:2318–2330 doi: 10.1109/TMI.2022.3161875.
- Grecco, Hernan E. 2018. “Pint.” https://pint.readthedocs.io/en/stable/
- Waskom M. seaborn: statistical data visualization. J. Open Source Softw. 2021;6:3021 doi: 10.21105/JOSS.03021.
Figures