2397
Magnetic Resonance Image Processing and Analysis Platform (MRI-PAP): A Windows-based Automated Pipeline for Multiple Sclerosis1Department of Neurology, University of Texas Health Sciences Center, Houston, TX, United States, 2Department of Cancer Systems Imaging, University of Texas MD Anderson Cancer Center, Houston, TX, United States, 3Department of Diagnostic and Interventional Imaging, University of Texas Health Sciences Center, Houston, TX, United States
Synopsis
Keywords: Software Tools, Data Processing, Pipeline
Neurologists and researchers rely on several software tools for neuroimaging data analysis. Limitations in these tools often necessitate the use of more than one program, spanning different coding languages and operating systems, which requires time, effort, and programming skills. MRI-PAP is a collection of pipelines with an easy-to-use graphical user interface (GUI) designed for MRI processing and analysis of multiple sclerosis patient images. These automated pipelines accept DICOM images as input and take them through a series of processing steps. MRI-PAP pipelines save time by accessing different packages from a single GUI and can be used without programming skills.
Introduction
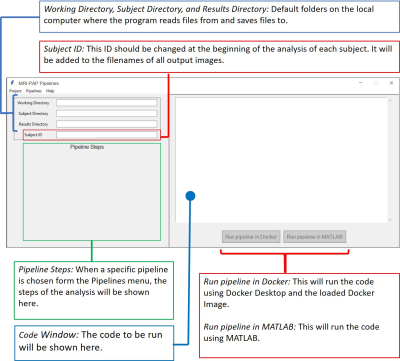
Many software packages for brain MRI data processing and analysis are available, such as FSL, FreeSurfer, ANTS, and SPM1-4. Each has their advantages and limitations. Therefore, neurologists and researchers often use more than one tool to carry out their imaging analysis. However, each package has its own syntax, interface, programming language, and operating system. Hence, it takes neurologists considerable time and effort to switch between tools. Nipype is an open-source neuroimaging Python package that provides interfaces to several of these existing packages and offers a Docker container that allows these packages to work on Windows5. However, programming knowledge is still required to use Nipype. Porcupine helps to reduce the coding burden by providing a graphical workflow with drag and drop interface, enabling researchers without extensive programming skills to design their own pipeline6. However, using Porcupine still requires some programming experience to build the workflow and achieve the desired analysis result. MRI-PAP solves this issue by providing a Windows-based, easy-to-use GUI that takes the data from raw DICOM files to processed, segmented, and analyzed Nifti images without the need for programming (Fig.1).Overview and Key Features
MRI-PAP is built for neurologists and requires no programming acumen. It is developed using Python 3, a free widely used programming language with a wide range of scientific computing libraries.MRI-PAP makes use of different programs/toolboxes to allow for straightforward processing and analysis of MR images, such as:
- The Docker image of the Nipype package5,7.
- Porcupine, a software package that automatically creates the Python code for a Nipype-based analysis pipeline6.
- Dcm2niix, a tool to convert imaging data from DICOM format to Nifti format8.
- SPM12 (Statistical Parametric Mapping), an open-source package that runs within the MATLAB environment4.
- SLF Lesion Filling, a MATLAB toolbox for brain MR Images9.
- Python libraries such as dicom2nifti and nibabel10,11.
MRI-PAP offers two modes of operation: single subject and batch processing.
Design and Implementation
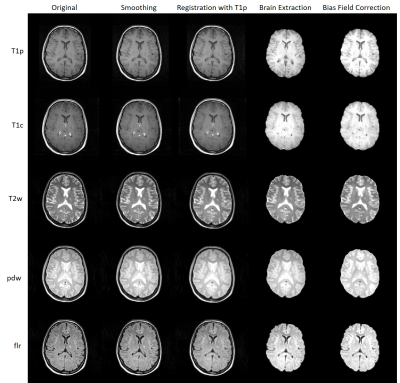
Fig. 2 shows a flowchart summarizing the different stages of the pipelines.Input data (DICOM format):
- Pre- and post-contrast T1-weighted images (T1p and T1c)
- A T2-weighted image (T2w)
- A proton density-weighted image (pdw)
- Fluid-attenuated inversion recovery (FLAIR) axial and sagittal images (flr and flr-sag)
Additional optional input data (DICOM format):
- Dynamic Susceptibility Contrast (DSC) MR perfusion images (DSC)
- Susceptibility Weighted Imaging (SWI) MR images (SWI)
Conversion pipeline:
- Convert input images from DICOM format to Nifti format.
- Check and fix flipped images. If the image is in RAS orientation, it will be flipped to LAS orientation.
Anatomical processing pipeline:
- Smoothing to reduce noise while preserving the underlying structure.
- Brain extraction using the T1p image.
- Registration of the T1c, T2w, pdw, flr, and flr-sag images to the T1p image.
- Application of brain mask to T1c, T2w, pdw, flr, and flr-sag images.
- Bias field correction to be applied on T1p, T1c, T2w, pdw, flr, and flr-sag images.
- Lesion filling on T1p, flr, and flr-sag images using lesion masks provided by the user.
Segmentation pipeline:
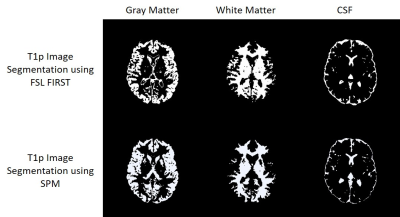
- Segmentation of T1p image into the different tissue classes; grey matter (GM), white matter (WM), and cerebrospinal fluid (CSF) using FSL FAST.
- Segmentation of T1p image into all the subcortical structures using FSL FIRST.
- Segmentation of T1p image into different tissue classes using SPM.
- Brain mask creation using the SPM segmentation output from the last step.
- Segmentation of T1p image into different tissue classes using SPM enhanced by the flr-sag image.
- Brain mask creation using the SPM segmentation output from the last step.
Dynamic Susceptibility Contrast (DSC) processing pipeline:
- Motion correction of the DSC image.
- Split the DSC image into individual volumes.
- Registration of the motion-corrected DSC image to the T1p image.
- Moving anatomical images, brain mask, and segmentation masks to the DSC space.
Susceptibility Weighted Imaging (SWI) processing pipeline:
- Merging of the original SWI echoes.
- Registration of the SWI echoes to the T1p image.
- Brain Mask application to the SWI echoes.
- Merging the registered and masked SWI echoes.
- Moving anatomical images, brain mask, and segmentation masks to the SWI space.
Usage Example
MRI-PAP was thoroughly tested and was applied for the analysis of several hundred images. Fig. 3 shows an example of the anatomical processing pipeline and Fig.4 shows an example of the segmentation output.Conclusion
MRI-PAP is a new software tool designed for neurologists to enable them to analyze their own imaging data without the need of switching between several software packages nor the burden of learning a programming language. MRI-PAP contains several processing and analysis pipelines that cover anatomical and structural images, as well as DSC and SWI image analysis that will be made publicly available via GitHub.Acknowledgements
Refaat Gabr is now with Biogen, Inc.References
1. Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. Fsl. Neuroimage. 2012 Aug 15;62(2):782-90.
2. Fischl B. FreeSurfer. Neuroimage. 2012 Aug 15;62(2):774-81.
3. Avants BB, Tustison N, Song G. Advanced normalization tools (ANTS). Insight j. 2009 Jun 4;2(365):1-35.
4. Ashburner J, Barnes G, Chen CC, Daunizeau J, Flandin G, Friston K, Kiebel S, Kilner J, Litvak V, Moran R, Penny W. SPM12 manual. Wellcome Trust Centre for Neuroimaging, London, UK. 2014 Jun 26;2464:4.
5. Gorgolewski K, Burns CD, Madison C, Clark D, Halchenko YO, Waskom ML, Ghosh SS. Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in python. Frontiers in neuroinformatics. 2011:13.
6. Van Mourik T, Snoek L, Knapen T, Norris DG. Porcupine: A visual pipeline tool for neuroimaging analysis. PLoS computational biology. 2018 May 10;14(5):e1006064.
7. Rad BB, Bhatti HJ, Ahmadi M. An introduction to docker and analysis of its performance. International Journal of Computer Science and Network Security (IJCSNS). 2017 Mar 1;17(3):228.
8. Li X, Morgan PS, Ashburner J, Smith J, Rorden C. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. Journal of neuroscience methods. 2016 May 1;264:47-56.
9. Valverde S, Oliver A, Lladó X. A white matter lesion-filling approach to improve brain tissue volume measurements. NeuroImage: Clinical. 2014 Jan 1;6:86-92.
10. Whitcher B, Whitcher MB. Package ‘oro. dicom’.
11. Brett M, Markiewicz CJ, Hanke M, Côté MA, Cipollini B, McCarthy P, Cheng CP, Halchenko YO, Cottaar M, Ghosh S. nipy/nibabel: 3.0. 0. Zenodo: Geneve, Switzerland. 2019 Dec.
Figures