2393
CMRsim - A Python package for MRI simulations incorporating complex organ motion and flow1University and ETH Zurich, Zurich, Switzerland
Synopsis
Keywords: Software Tools, Simulations, Cardiovascular
We present CMRsim, a new Python package for MR simulations efficiently incorporating complex organ motion and flow. The MR signal can be calculated using both Bloch simulations as well as analytical signal models. The package leverages TensorFlow2 for GPU acceleration and thereby facilitates fast simulations while also providing a compilation-free framework for prototyping and community contributions. Significant effort has been put on maintainability and software engineering best practices. The API reference as well as information on installation and how to get started is publicly available at: https://people.ee.ethz.ch/~jweine/cmrsim/latest/index.html
Keywords: Open-source, Simulation, Cardiovascular, Motion, Reproducible science, Digital twin
Introduction
Simulations and in-silico analysis are becoming increasingly relevant in the development of optimal imaging techniques in order to improve and optimize various parts of the imaging pipeline1-4 or to demonstrate the ability to discover pulse sequences using iterative learning5.One area where sequence optimization could yield significant improvements is cardiovascular MR (CMR). The deformation of the heart during the cardiac and respiratory cycles is non-rigid, prompting advanced simulation solutions. Likewise, cardiovascular flows can be transient and even turbulent resulting in highly complex motion of magnetization in blood. While there are many existing simulations packages6-9, they either assume spatially static systems or are not suited for incorporating motion typical for CMR. Here we present a newly developed MR simulation framework, CMRsim, which incorporates complex motion of large numbers of material points.
Design and Features
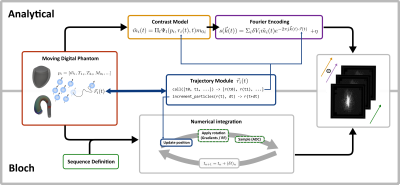
CMRsim is written in Python, leveraging the language's vast collection of available packages and easily understandable style to boost the open-source nature of the project. The computationally demanding operations are performed using TensorFlow2, which provides efficient GPU acceleration as well as compatibility for systems without GPUs. Emphasis was put on design to ensure CMRsim complies with open-source tool standards including best practices for coding style, documentation and continuous development.Depending on the application, the MR signal is either calculated numerically by solving the Bloch equation or by applying analytical operators. While Bloch simulations require fewer assumptions, evaluating analytical operators usually is significantly less computationally intensive. As such, the presented package contains a module for each approach which will be subsequently referred to as analytical and Bloch simulation. A conceptual overview of both simulation types is given in Figure 1.
Analytic Simulations
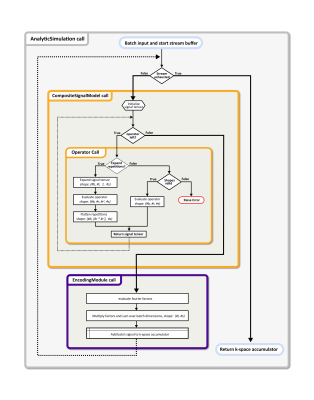
Analytical simulations are implemented in a pipe-and-filter pattern, where the operators acting on the apparent transverse magnetization can be composed into a forward model. The analytical simulation module already provides a set of common operators (see the API reference for a complete list), while the software structure allows adding module prototypes without integration into the package. Additionally, the modules reduce redundant computations on evaluating the signal model. Fourier encoding is performed by dedicated encoding modules, supporting multiple interfaces to define the corresponding k-space trajectory (see API reference). The input object is streamed to the GPU in batches using the TensorFlow data API, and accumulating the k-space signal is managed by a composite simulation module containing the forward model, the encoding and an optional reconstruction module. Furthermore, additional functionality such as summarization or saving the configured simulation is provided. A flow diagram for analytical simulation evaluation with CMRsim is presented in Figure 2.
Bloch Simulations
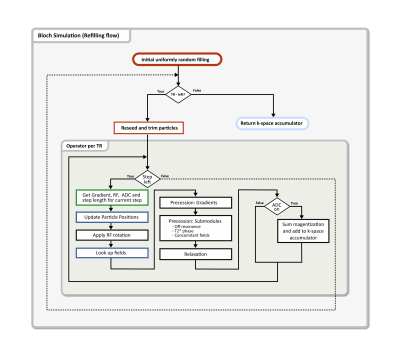
Bloch simulations are used to numerically solve the Bloch equation for a large number of particles (isochromats). To include motion into Bloch simulations, the particle/isochromat positions must be updated in each numerical integration step and the temporal step size must be sufficiently small to minimize errors i.e. typically below 10us. Saving the position of many millions of particles over durations of ~100ms prior to simulation becomes infeasible due to exploding file sizes. To address this issue, CMRsim offers an efficient and flexible solution to incorporate motion evaluation into the integration loop, thereby facilitating large scale flow simulations. Simulations are performed by first instantiating a module that receives gradient and RF waveforms as well as ADC events defined as numpy arrays. Calling the module requires an instance of a trajectory module to be passed, which handles particle position updates using Tensorflow2-graph compatible code. A flow chart of the simulation procedure is given in Figure 3. A collection of trajectory modules are already contained in the package. A GPU based implementation of nearest neighbor and trilinear interpolation on 3D+t+N dimensional look-up maps is available and serves as base implementation for trajectory modules.
Examples and Performance
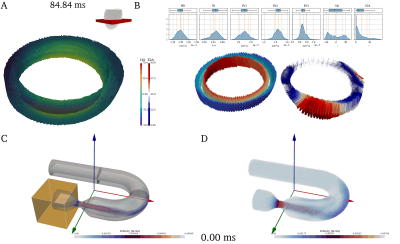
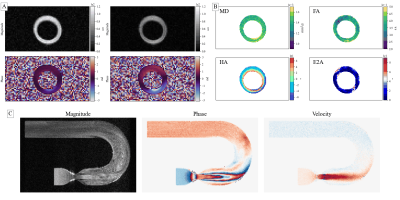
Example 1 - To demonstrate simulations, a cardiac diffusion weighted spin-echo experiment with 8 coils and 13 diffusion directions at 6 trigger delays is presented (~70k particles / 624 images). Figure 4a shows the input mesh and its motion. The resulting images are shown in Figure 5a. The simulation took ~6 minutes on a NVIDIA Titan RTX 24Gb.Example 2 - As a demonstration for Bloch simulations a velocity encoded spoiled GRE with laminar flow within a stenotic U-bend was performed. To address in- and outflow, reseeding provided by CMRsim was used, which estimates the particle density before every excitation pulse and fills voids within the user-defined inflow region. As the implementation uses the pyvista10 package, meshes in the vtk format are supported. The input including refilling is illustrated in Figure 4b and the resulting images can be found Figure 5b. For a 350ms scan with a mean particle density of 40/mm3 the simulation took ~80 minutes on a NVIDIA Titan RTX 24Gb.
Conclusion
CMRsim offers a simulation framework that incorporates complex motion and flow. The software structure as well as the infrastructure supports usability and reproducibility of simulation experiments.Code: https://gitlab.ethz.ch/jweine/cmrsim
Documentation: https://people.ee.ethz.ch/~jweine/cmrsim/latest/index.html
Acknowledgements
References
1. Xanthis CG, Bidhult S, Kantasis G, Heiberg E, Arheden H, Aletras AH. Parallel simulations for QUAntifying RElaxation magnetic resonance constants (SQUAREMR): An example towards accurate MOLLI T1 measurements. J. Cardiovasc. Magn. Reson. 2015;17:1–15 doi: 10.1186/s12968-015-0206-1.
2. Ma D, Gulani V, Seiberlich N, et al. Magnetic resonance fingerprinting. Nature 2013;495:187–192 doi: 10.1038/nature11971.
3. Weiss, Tomer, et al. "PILOT: Physics-informed learned optimal trajectories for accelerated MRI." arXiv preprint arXiv:1909.05773 2.11 (2019).
4. Zhu, Bo, et al. "AUTOmated pulse SEQuence generation (AUTOSEQ) using Bayesian reinforcement learning in an MRI physics simulation environment." Proceedings of the 26th Scientific Meeting, International Society for Magnetic Resonance in Medicine. 2018.
5. Loktyushin A, Herz K, Dang N, et al. MRzero - Automated discovery of MRI sequences using supervised learning. Magn. Reson. Med. 2021;86:709–724 doi: 10.1002/MRM.28727.
6. Stöcker T, Vahedipour K, Pflugfelder D, Shah NJ. High-performance computing MRI simulations. Magn. Reson. Med. 2010;64:186–193 doi: 10.1002/MRM.22406.
7. Xanthis CG, Venetis IE, Chalkias A V., Aletras AH. MRISIMUL: A GPU-based parallel approach to MRI simulations. IEEE Trans. Med. Imaging 2014;33:607–617 doi: 10.1109/TMI.2013.2292119.
8. Liu F, Velikina J V., Block WF, Kijowski R, Samsonov AA. Fast Realistic MRI Simulations Based on Generalized Multi-Pool Exchange Tissue Model. IEEE Trans. Med. maging 2017;36:527–537 doi: 10.1109/TMI.2016.2620961.
9. Jochimsen TH, Von Mengershausen M. ODIN-Object-oriented development interface for NMR. doi: 10.1016/j.jmr.2004.05.021.
10 Sullivan CB, Kaszynski AA. PyVista: 3D plotting and mesh analysis through a streamlined interface for the Visualization Toolkit (VTK). J. Open Source Softw. 2019;4:1450 doi: 10.21105/JOSS.01450.
Figures