2392
NiGSP: Graph Signal Processing on multimodal MRI data.1Neuro-X Institute, Ecole polytechnique fédérale de Lausanne, Geneva, Switzerland, 2Department of Radiology and Medical Informatics (DRIM), Faculty of Medicine, University of Geneva, Geneva, Switzerland, 3Neuro-X Institute, EPFL Valais, Clinique Romande de Réadaptation, Sion, Switzerland, 4Department of Clinical Neurosciences, Geneva University Hospital (HUG), Geneva, Switzerland
Synopsis
Keywords: Software Tools, Brain Connectivity
NiGSP is a python-based toolbox aimed at facilitating the adoption of graph signal processing with an emphasis on multimodal brain imaging data. We present its standard workflow, that allows basic filtering operations and metrics computations, and we introduce a novel application to cerebral stroke consisting in the creation of a subject-specific anatomical lesion-based filter to be applied on functional MRI timeseries.Introduction
Graph Signal Processing (GSP) is a novel method to process temporal signals, taking into account the features of the graph they are embedded in. It provides an integrated framework to approach multimodal MRI, mainly used for functional MRI (fMRI) fluctuations embedded in the graph of structural connectivity (SC)1–6. For instance, it has been used to elucidate pathological processes affecting brain function and structure in various clinical populations4. Retrieving fMRI fluctuations coupled or uncoupled with the structural connectivity at a coarse resolution revealed patterns of function-structure alignment that agree with cognitive domain hierarchy and genetic preferential expression5. Such coupling has been shown to uniquely characterise individuals7, and to change between healthy subjects and clinical populations2.Here we present NiGSP8, an open-development python-based toolbox aimed at facilitating the adoption of GSP, with an emphasis on multimodal brain imaging and clinical application. It offers a set of functions for GSP filter creations, filtering, visualisation, and metric computations, together with a standard workflow accessible through command line interface to compute such metrics automatically.
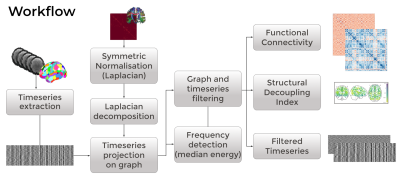
Workflow
NiGSP workflow implements the GSP workflow detailed in 5,7 to assess the alignment of time-varying fMRI signals to the underlying SC graph. Briefly, after construction and eigendecomposition of the graph Laplacian, it projects the regional timeseries extracted at the same nodes into the graph Fourier space (graph Fourier transform). Graph spectral filtering is then implemented to divide the fMRI timeseries in parts with equal spectral energy. Beside simple filtering, the toolbox allows to compute GSP-derived metrics such as structural decoupling index5 and filtered Functional Connectivity (FC)3, to compute data surrogates for statistical thresholding, and to visualise results in matricial or graph form (see Figure 1). Compliance with different data format (3D-4D NIfTI, .txt-.mat files) is implemented.Novel Application
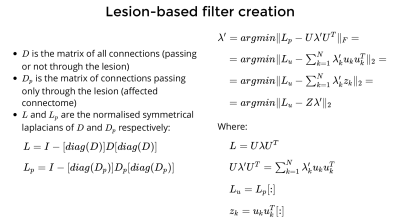
NiGSP can be used as a library for advanced and novel applications. We introduce here a possible application to cerebral stroke, with the creation of filters based on graph disruption, reproducing brain lesions impacting structural connectivity. This is done by computing a set of weights that minimises the difference between the Laplacian normalisations of a whole brain graph and its subgraph of connections passing only through the lesion, called affected connectome9 (see Figure 2). This set of weights is then used to filter the timeseries through graph Fourier transform, aiming at projecting the effect of the anatomical lesion on whole-brain FC.Methods
We selected three subjects, with lesions differing in size and location (see Figure 3a), from a cohort of 44 ischaemic stroke patients undergoing MRI scanning (3T Magnetom Prisma, Siemens) during the first week post-stroke (5.2 ± 2.6 days). Written informed consent was obtained from each participant and the study was authorised by the local ethical committee.Diffusion-weighted images (pulsed gradient spin echo, TR=5000ms; TE=77ms; slices=84; 1.6×1.6×1.6mm3 resolution; FoV=234×234mm2; readout=1630Hz/pixels; 64-channel head coil; acceleration factor=3), including volumes in opposite phase encoded direction without diffusion weighting, were preprocessed following 9. T1-weighted images (MPRAGE, TR=2300ms; TE=2.96ms; flip angle=9°; slices=192; 1×1×1mm3 resolution, FoV=256×256mm2) were used to hand-draw lesion masks and preprocessed following 9. Resting-state BOLD data (echo-planar imaging, 385 volumes; TR=1s; TE=32ms; flip angle=58°; slices=75; 2×2×2mm3 resolution; FoV=224×224mm2; acceleration factor=5) were skull stripped, motion realigned, bias field corrected, nuisance regressed (motion parameters, 1st derivatives, and first 5 Legendre polynomials), and smoothed (4mm FWHM). SC was obtained using a modified version of the Destrieux atlas (detailed in 9). The same atlas was used to extract the average BOLD timeseries for each parcel, and these were filtered using a subject-specific lesion-based filter created following the steps above. FC and “global” FC (GFC, expressed as FC node strength) were computed for original and filtered timeseries and compared with each others.
Results
NiGSP is available on GitHub (https://github.com/miplabch/nigsp), it is packaged and released under Apache-2.0 licence on pypi (https://pypi.org/project/nigsp/) or as ready-to-use docker container. Development is ongoing, and all contributions are welcomed and recognised following the all-contributors specification.The three subjects analysed here presented lesions with different size and location (see Figure 3a). The created filters (Figure 3b) resulted in attenuated low frequencies and augmented high frequencies, with increasing extremization of weights related to lesion size.
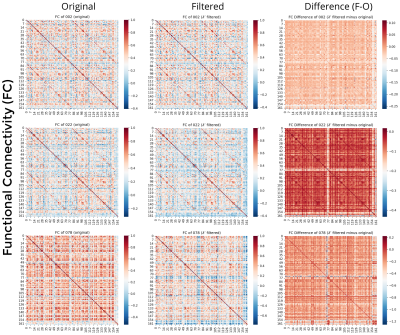
Comparing FC matrices (Figure 4), subject 2 presented generally modest differences, with the exception of the right central sulcus (in correspondence of the lesion), capsula interna, and pons. Subject 22 presented of distant regionsFC decrease mainly in cerebellum, capsula interna, and pons, suggesting disrupted long range FC of distant regions. Subject 78 presented FC decreases in left and right temporal lobe, and right occipital and parietal cortices.
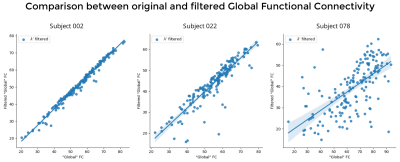
Comparing GFC (Figure 5), we see decreasing similarity between original and filtered signals and increasing plot dispersion with increasing lesion size.
Conclusion
We introduced the toolbox NiGSP, implementing the GSP framework for neuroimaging in a user-friendly and generalisable fashion.Overall, GSP and NiGSP offer a new promising framework for multimodal MRI integration and clinical interpretation. Future studies will allow to further elucidate the clinical value of the proposed lesion filter application.
Acknowledgements
No acknowledgement found.References
1. Abramian D, Larsson M, Eklund A, Aganj I, Westin CF, Behjat H. Diffusion-informed spatial smoothing of fMRI data in white matter using spectral graph filters. Neuroimage. 2021;237(April):118095. doi:10.1016/j.neuroimage.2021.118095
2. Bortolin K, Delavari F, Preti MG, et al. Neural substrates of psychosis revealed by altered dependencies between brain activity and white-matter architecture in individuals with 22q11 deletion syndrome. NeuroImage Clin. 2022;35(May):103075. doi:10.1016/j.nicl.2022.103075
3. Griffa A, Amico E, Liégeois R, Van De Ville D, Preti MG. Brain structure-function coupling provides signatures for task decoding and individual fingerprinting. bioRxiv. 2021. doi:10.1101/2021.04.19.440314
4. Kong Z, Sun L, Peng H, Zhan L, Chen Y, He L. Multiplex Graph Networks for Multimodal Brain Network Analysis. arXiv. July 2021. doi:10.48550/arXiv.2108.00158
5. Preti MG, Van De Ville D. Decoupling of brain function from structure reveals regional behavioral specialization in humans. Nat Commun. 2019;10(1):1-7. doi:10.1038/s41467-019-12765-7
6. Tarun A, Behjat H, Bolton T, Abramian D, Van De Ville D. Structural mediation of human brain activity revealed by white-matter interpolation of fMRI. Neuroimage. 2020;213(November 2019):116718. doi:10.1016/j.neuroimage.2020.116718
7. Griffa A, Amico E, Liégeois R, Van De Ville D, Preti MG. Brain structure-function coupling provides signatures for task decoding and individual fingerprinting. Neuroimage. 2022;250(November 2021):118970. doi:10.1016/j.neuroimage.2022.118970
8. Moia S. NiGSP: A python library (and toolbox!) to run Graph Signal Processing on multimodal MRI data. Zenodo. July 2022. doi:10.5281/zenodo.6373436
9. Egger P, Evangelista GG, Koch PJ, et al. Disconnectomics of the Rich Club Impacts Motor Recovery after Stroke. Stroke. 2021;(June):2115-2124. doi:10.1161/STROKEAHA.120.031541
Figures