2389
FGPA based accelerator for pMRI using coil compression1Electrical and Computer Engineering Department, Comsats University Islamabd, Islamabad, Pakistan, 2Electrical and Computer Engineering Department, Comsats University Islamabad, Islamabad, Pakistan, 3Comsats University Islamabad, Islamabad, Pakistan
Synopsis
Keywords: Parallel Imaging, Cardiovascular
Parallel MRI (e.g., SENSE, GRAPPA) accelerates the data acquisition in modern clinical scanners using multiple receiver coils. However, it results in more computational demands on general purpose computers. Coil compression is a promising way to address the computational cost and memory requirements associated with a large number of receiver coils. In this work, a novel FPGA based hardware accelerator is designed to perform coil compression using QR decomposition. In-vivo reconstruction results from 30 coil cardiac data set, show that the proposed accelerator elevates the speed and memory constraints while preserving the image quality.
Introduction
Parallel MRI (pMRI) methods allow faster data acquisitions using multiple receiver coils. However, data processing using multiplier receiver coils causes exponential growth in the reconstruction time of pMRI methods such as sensitivity encoding (SENSE)1 and Generalize Auto calibrating Partial Parallel Acquisition (GRAPPA)2. Recently, Eigen value methods such as principal component analysis (PCA)3 have been adopted to compress the multiple channel information into fewer receiver coils data while retaining the important information, thus significantly reducing computer memory requirements and reconstruction time for pMRI methods.Singular Value Decomposition (SVD) of a matrix is a useful tool for dimension reduction using PCA4. SVD5 decomposes a given matrix (Anxm) into three main components i.e. $$$A = U S V^T$$$ where, S is singular diagonal matrix containing eigen values; U and V are columns and rows orthogonal matrices containing the information of Eigen vectors. The time complexity of computing SVD of an n×m matrix is O(nm min(m, n))6;and for high rank data sets, SVD becomes computationally expensive. Moreover, SVD does not have independent operations which could be exploited using hardware or software solutions to speed the process of compression. In this work, an FPGA based accelerator for coil compression is designed which employs QR decomposition using modified Gram-Schmidt7 to obtain accurate information of the Eigen vectors required for data compression. QR decomposition8 procedure decomposes a given matrix (Anxm) into two main components i.e. $$$A = QR$$$ where, Q is the orthogonal matrix and R is the upper triangular matrix. In modified Gram-Schmidt method, all the column vectors of matrix (Anxm) are modified to be orthogonal to the first column vector of marix (Anxm)for faster computation as it orthogonalizes the column vectors simultaneously9. The proposed accelerator exploits the inherent parallelism in QR decomposition and works in coordination with an on-chip processor to perform image reconstruction using multiple receiver coil data. It is demonstrated that coil compression using the proposed accelerator can be highly effective for GRAPPA and SENSE methods in terms of reconstruction time and quality.
Method
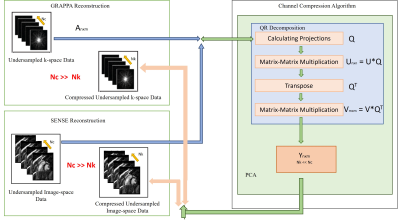
In this work, an FPGA based accelerator for channel compression is proposed to accelerate GRAPPA and SENSE reconstruction without compromising the image quality as shown in Fig. 3. The proposed accelerator obtains accurate information of Eigen vectors based on QR decomposition using modified Gram–Schmidt method which decomposes the given matrix (Anxm) into an orthogonal matrix(Qnxm) and an upper triangular matrix (Rmxm)10; the pseudo code is shown in Fig. 1.Rmxm and (Rmxm)T compute the orthonormal matrices (Unxn) and (Vmxm) by orthogonalizing all the column vectors with respect to the first column vectors of each matrix. Unxn and Vmxm are columns and rows of Eigen vectors to calculate the Eigen values of receiver coil data matrices. Eigen vectors calculate the dimensionally reduced matrix contains the essential and compressed information with fewer coils as shown in Fig. 2. The reduced coil data from channel compression algorithm works in coordination with an on-chip ARM processor11 using AXI4-stream-interconnect12 for efficient GRAPPA and SENSE reconstruction.
For implementation of the proposed accelerator, High-Level Synthesis(HLS) framework provided by VIVADO HLS tool13 is used. HLS framework provides flexibility in terms of architectural optimization for different configuration settings(SENSE and GRAPPA). The design flow of the proposed accelerator is:
(i) Generation of high precision source code of the coil compression algorithm bases on C/C++ language using floating-point data type in compliance with VIVADO HLS design constraints. As a result, VIVADO HLS tool automatically generates the RTL code of the 32-bit single-precision floating-point FPGA-based accelerator for channel compression.
(ii)The optimization pragmas such as “HLS PIPELINE”(pipelines the data processing) and “HLS UNROLL"(unrolls loops to provide data level parallelism in algorithm) are used to generate RTL. RTL is simulated and verified through C/C++ test bench based RTL wrappers. Finally, RTL design of the channel compression accelerator consisting of IO ports is packaged as HLS intellectual property(IP) block referred as “Channel Compression Accelerator”. The proposed IP is well equipped with the required interfaces to work in coordination with on-chip ARM processor where GRAPPA and SENSE reconstructions are performed on the compressed data.
(iii) Finally, the bit file of 32-bit floating-point Channel Compression Accelerator is generated for implementation on Xilinx FPGA development board ZCU102 that contains an on-chip ARM Cortex®-A53 64-bit quad-core processor.
Result and Discussion
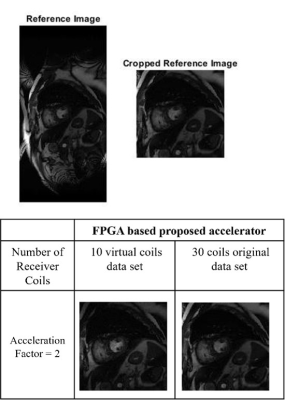
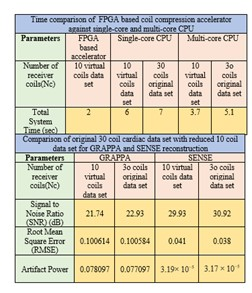
The experiments are performed on a 30 channel cardiac data set using various configuration settings as shown in Table 1. For a fair comparison, the performance of GRAPPA and SENSE reconstructions with FPGA based channel compression is evaluated against single-core and multi-core CPU. The results show that the proposed accelerator speeds up the reconstruction times (GRAPPA and SENSE) up to 15x with acceptable SNR as shown in Table 1. The proposed accelerator utilizes less resources to reduce receiver coil data and reduces computer memory requirements without degrading image quality as shown in Fig. 3.Conclusion
The proposed work presents a novel FPGA based accelerator for channel compression which exploits the inherent parallelism in the QR decomposition. The results show that coil compression using the proposed accelerator offers speed up in the reconstruction time with for GRAPPA and SENSE methods with no compromise on image quality.Acknowledgements
References
[1] SENSE: Sensitivity encoding for fast MRI. (1999, October 28). WILEY., from https://onlinelibrary.wiley.com/doi/abs/10.1002/(SICI)1522-2594(199911)42:5%3C952::AID-MRM16%3E3.0.CO;2-S
[2] GPU accelerated Cartesian GRAPPA reconstruction using CUDA. (2022, February 24). GPU Accelerated Cartesian GRAPPA Reconstruction Using CUDA - ScienceDirect. https://doi.org/10.1016/j.jmr.2022.107175
[3]Zwart, N. R. (n.d.). (ISMRM 2021) PCA and U-Net based Channel Compression for Fast MR Image Reconstruction. (ISMRM 2021) PCA and U-Net Based Channel Compression for Fast MR Image Reconstruction. https://archive.ismrm.org/2021/1752.html
[4] How Are Principal Component Analysis and Singular Value Decomposition Related? (n.d.). How Are Principal Component Analysis and Singular Value Decomposition Related?, from https://intoli.com/blog/pca-and-svd/
[5] QR-decomposition based SENSE reconstruction using parallel architecture. (2018, February 2). QR-decomposition Based SENSE Reconstruction Using Parallel Architecture - ScienceDirect. from https://www.sciencedirect.com/science/article/pii/S0010482518300210?casa_token=gr_yvACas_sAAAAA:kdPU07gDSQMNMlJLV5VCOx_rHprE5BU1PMO3kKFTJu8k3cwsQh8mvj2pytfByaITqjExoTgBsw
[6] Time Complexity of Singular Value Decomposition. (2015, March 23). Stack Overflow. from https://stackoverflow.com/questions/29218823/time-complexity-of-singular-value-decomposition
[7]Classical Gram–Schmidt vs Modified Gram–Schmidt. (n.d.). from https://arnold.hosted.uark.edu/NLA/Pages/CGSMGS.pdf
[8] Sharma, A., Paliwal, K. K., Imoto, S., & Miyano, S. (2012, September 25). Principal component analysis using QR decomposition - International Journal of Machine Learning and Cybernetics. SpringerLink. from https://link.springer.com/article/10.1007/s13042-012-0131-7
[9] Lecture23. (n.d.). Lecture23., from https://www.math.uci.edu/~ttrogdon/105A/html/Lecture23.html
[10] ACM Digital Library. (n.d.). ACM Digital Library, from https://dl.acm.org/doi/abs/10.1145/1513895.1513904?casa_token=Xzzy94T_PaQAAAAA:H-il4zC__xV_MNXqwfhLMZ3ec3jJ5ufWripAydn_uPACBe9UESaF02x0sIOFkdzgWAWy4yldC5u0
[11]What is an ARM processor? (2022, July 21). What Is an ARM Processor ?, from https://www.redhat.com/en/topics/linux/what-is-arm-processor
[12] AXI4-Stream Interconnect. (n.d.). Xilinx., from https://www.xilinx.com/products/intellectual-property/axi4-stream_interconnect.html
[13] Xilinx Vivado High Level Synthesis: Case studies. (n.d.). Google Scholar, from https://scholar.google.com/scholar_lookup?title=Xilinx%20Vivado%20High%20Level%20Synthesis&publication_year=2014&author=D.%20O%27Loughlin&author=A.%20Coffey&author=F.%20Callaly&author=D.%20Lyons&author=F.%20Morgan
Figures