2383
Double-echo phase-incrementing SSel-MQC (pi-SSelMQC) in biomarker imaging with full signal recovery and excellent lipid and water suppression
Qiuhong He1, Hong Yuan2,3, and Yen-Yu Ian Shih2,4,5
1The School of Health Sciences, Purdue University, West Lafayette, IN, United States, 2Biomedical Research Imaging Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States, 3Radiology, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States, 4Neurology, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States, 5Center for Animal MRI, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
1The School of Health Sciences, Purdue University, West Lafayette, IN, United States, 2Biomedical Research Imaging Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States, 3Radiology, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States, 4Neurology, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States, 5Center for Animal MRI, University of North Carolina at Chapel Hill, Chapel Hill, NC, United States
Synopsis
Keywords: Pulse Sequence Design, Spectroscopy
A double-echo pi-SSelMQC method was developed to recover 100% biomarker signals, as compared to spin-echo pulse sequences, with excellent water and lipid suppression. Signals from both ZQ→DQ and DQ→ZQ coherence transfer pathways were detected. By synchronization of phase-encoding steps and RF phase increments of either MQ-preparation or MQ-transfer pulse, the biomarker images from different MQ-pathways were resolved and shifted away from residual lipid and water signals. Full lactate and polyunsaturated fatty acids (PUFA) signals were recovered in pi-SSelMQC imaging using yogurt phantoms, vegetable oil, and murine 344SQ lung tumors grown subcutaneously on the right thigh of syngeneic 129X1/SvJ male mice.Introduction
The Selective Multiple-Quantum Coherence transfer (Sel-MQC) methods enable in vivo mapping of biomarker distributions in biological tissues containing high-fat content with complete lipid and water suppression in a single-scan [1]. The method was employed in proton detection of the antineoplastic agent Iproplatin in a mouse tumor model [2] and polyunsaturated fatty acids (PUFA) in human breast cancer [3, 4]. However, only one of the ZQ→DQ and DQ→ZQ coherence transfer pathways was detected in order to suppress lipid and water. Half of the biomarker signals was lost as compared to the spin echo pulse sequence. In this report, we present a novel phase-incrementing soft Sel-MQC (pi-SSelMQC) method for simultaneous detection of both ZQ→DQ and DQ→ZQ pathways with full biomarker signal recovery. We have developed the double-echo pi-SSelMQC pulse sequences to achieve excellent lipid and water suppression. The methods are applicable for fast MRSI in extracranial tissues.Methods
The soft Sel-MQC (SSel-MQC) method [1] was modified into the pi-SSelMQC sequence by incrementing the RF phase of the second or the last 90° pulse for MQ-coherence preparation or transfer, respectively, in synchronization with the phase-encoding gradient (Fig. 1). Since the coherence order of the DQ→ZQ and ZQ→DQ coherence transfer pathways have opposite signs, this procedure introduces opposite image offsets of signals from the two different MQ-coherence transfer pathways, away from unwanted spurious signals of residual water and lipids. The method enables simultaneous detection of both DQ→ZQ and ZQ→DQ pathways for a full recovery of the biomarker signals. Acquiring both pathways in the same acquisition window, however, reintroduced lipid and water signals (Fig. 2a). To address the issue, we have designed the double-echo pi-SSelMQC pulse sequences (Fig. 3) to acquire two separate MQ-coherence transfer echoes from the DQ→ZQ and ZQ→DQ coherence transfer pathways. We have previously observed that the two MQ-transfer echoes in the Sel-MQC experiments were separated by 2t1 [1], permitting insertion of gradient pulses between the two MQ-coherence transfer echoes to suppress unwanted signals (Fig. 3). The biomarker signals from the two MQ-coherence pathways can be digitally added for full recovery without 50% loss of the biomarker signal. The double-echo pi-SSelMQC achieved excellent lipid and water suppression, similar to the original SSel-MQC experiments.Results
The pi-SSelMQC pulse sequences were implemented on a Bruker 9.4T BioSpec 94/30USR AVII MRI spectrometer. The system has the BFG-240/120-S13B shielded gradients (12cm bore size) with Integrated Shims (Resonance Research, Inc) and the maximum gradient strengths of 999.63, 1,001.9, and 1,001.6 mT/m respectively in x-, y- and z-directions. We detected yogurt lactate signals from both DQ→ZQ and ZQ→DQ coherence transfer pathways in two-dimensional pi-SSelMQC imaging experiments using a gap resonator RF coil (data will be presented). In one-dimensional (1D) CSI experiments using a single-loop RF coil, we have observed tumor lactate signals in vivo in a mouse lung cancer model simultaneously from both DQ→ZQ and ZQ→DQ coherence pathways (Fig. 2a), and separately from either DQ→ZQ (Fig. 2b) or ZQ→DQ (Fig. 2c) coherence pathway. The mouse tumor was grown into 10.73 mm x 10.72 mm x 8.63 mm size after inoculating mouse lung cancer 344SQ cells (0.5 million in 100 ul of serum-free medium) subcutaneously in the right thigh region of the syngeneic 129X1/SvJ male mice. When both MQ-coherence transfer pathways were detected in the single acquisition window, intense lipid and water signals were reintroduced with gradient ratio g1:g2:g3 = 1: -1: 2 [Fig. 2a]. When using two acquisition windows separately for the two MQ-coherence transfer pathways, excellent lipid and water suppression were achieved for fast imaging of PUFA (Fig. 4) or lactate (Fig. 5) in the presence of read gradient. Fast 3D imaging of these biomarkers were also demonstrated using the double-echo pi-SSelMQC techniques.Conclusion
When synchronizing RF phase-incrementation and phase-encoding gradients, we have recovered full biomarker signals in pi-SSelMQC imaging in phantoms and in vivo in a mouse tumor model. The fast double-echo pi-SSelMQC imaging methods offer outstanding lipid and water suppression in a single-scan with 100% biomarker signal recovery.Acknowledgements
We thank the funding support from Linebarger Comprehensive Center (P30 CA016086), Bowles Center for Alcohol Studies (P60 AA011605), and Carolina Institute for Developmental Disabilities (U54 HD079124) for Center for Animal MRI facilities at the University of North Carolina at Chapel Hill.References
1. He, Q., et al., Single-scan in Vivo Lactate Editing With Complete Lipid and Water Suppression by Selective Multiple-Quantum-Coherence Transfer (Sel-MQC) With Application to Tumors. J. Magn. Reson. B, 1995. 106(3): p. 203-11. 2. He, Q., et al., Proton observation of the antineoplastic agent iproplatin in vivo by selective multiple quantum coherence transfer (Sel-MQC). Magn. Reson. Med., 1995. 33(3): p. 414-416. 3. He, Q., et al., In Vivo MR Spectroscopic Imaging of Polyunsaturated Fatty Acids (PUFA) in Healthy and Cancerous Breast Tissues by Selective Multiple-Quantum Coherence Transfer (Sel-MQC): A Preliminary Study. Magn. Reson. Med., 2007. 58: p. 1079-1085. 4. Zhu, H., D. Rubin, and Q. He, The Fast Spiral-SelMQC Technique for In Vivo MR Spectroscopic Imaging of Polyunsaturated Fatty Acids in Human Breast Tissue. Magn. Reson. Med., 2012. 67: p. 8–19.Figures
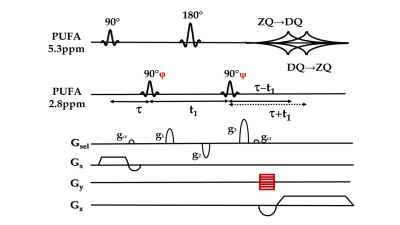
Fig. 1 The pi-SSelMQC
pulse sequence with RF phase increments of the second 90° pulse (or
alternatively the last 90° pulse) in synchronization with the spatial
phase-encoding steps. The MQ-selection
gradient g1, g2 and g3 select MQ-coherence transfer pathways. The gcr symbolizes the TE-crusher gradient. For fast biomarker imaging, a frequency-encoding gradient was applied replacing one of the phase-encoding dimensions in the Cartesian chemical shift imaging (CSI) schedule of 2D multi-voxel MRSI.
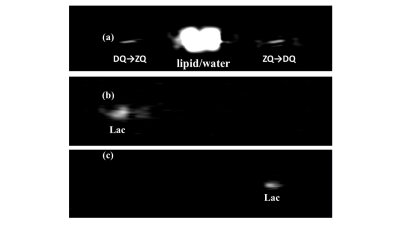
Fig. 2 The 1D pi-SSelMQC
CSI measurement of the lactate signals from 344SQ murine lung cancer growing in
the right thigh of the nude mouse from (a) both MQ coherence pathways or (b)
DQ→ZQ and (c) ZQ→DQ pathways separately with g1:g2:g3
= 1:-1:2, 1:0:2, and 0:-1:2, respectively.
The tumor size was 10.73 mm x 10.72 mm x 8.63 mm.
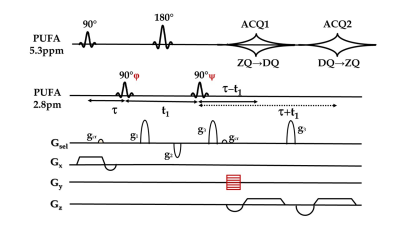
Fig. 3 A double-echo pi-SSelMQC
pulse sequence to detect the two MQ-coherence transfer echoes of PUFA. The same sequence was also applied in lactate
imaging using coupled spins at 1.3ppm and 4.2ppm.

Fig. 4
The PUFA images acquired separately from the two
MQ-coherence transfer echoes in the soybean oil phantom using a Doty 25
mm volume coil in an axial slice using the double-echo pi-SSelMQC pulse
sequence.
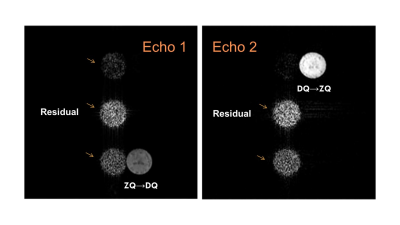
Fig. 5 Lactate images from two MQ-coherence transfer echoes using the
double-echo pi-SSelMQC pulse sequence to recover 100% lactate in a
yogurt phantom using a Doty 25 mm volume coil. Residual lipid and water are separated from the
biomarker images. Note that complete lipid
and water suppression is achievable with stronger MQ-coherence selection gradients.
DOI: https://doi.org/10.58530/2023/2383