2370
Anatomical priors informed tractography. Evaluation on the DiSCo synthetic dataset1Univ Rennes, INRIA, CNRS, INSERM, IRISA UMR 6074, Empenn ERL U-1228, F-35000, Rennes, France, 2Signal Processing Laboratory (LTS5), Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland, 3Department of Computer Science, Université de Sherbrooke, Sherbrooke, QC, Canada
Synopsis
Keywords: Data Processing, Tractography & Fibre Modelling
White matter fiber tracking using diffusion magnetic resonance imaging (DWI) provides a powerful approach to map brain connections, but they are not completely reliable both in academic and clinical context. This is because most methods infer global connectivity from local information,leading to a high rate of false positive. We propose a method to include anatomical prior in the tractography algorithm. We show on synthetic DiSCo data that including this prior on two state-of-the-art tractography algorithms could improve the overall shape of the tractograms.Introduction
White matter fiber tracking using diffusion magnetic resonance imaging provides a non invasive powerful approach to map brain connections. However, for now, those methods still lack reliability both in clinical and academic context 1, 2. In 3, it was shown that most of the actual state-of-the-art algorithms, although succeeding to extract most of the fibers bundles, also estimate many false bundles, i.e. bundles that do not exist in the brain, leading to significant false positive rate.This may come from several different sources but one of the most important is the fact that, in tractography, the overall goal is to infer a global structure from local information that causes the algorithm to take, in complex regions, non-realistic decisions. To overpass this limit, it has been proposed in 4 to add global information to the algorithms in the form of anatomical priors to guide the tractography to increase the quality of the tractograms.In this work, we propose a new method, based on 4, of creation and combination of anatomical priors and Fibers Orientation Distribution (FOD) model 5. We then evaluate this method on the synthetic DiSCo dataset 6, showing the influence of various parameters on the quality of the tractograms obtained with different state-of-the-art tractography algorithms, informed by anatomical priors.
Materials
The data used in this work come from the DiSCo challenge numerical phantoms 6 .The anatomical priors were generated from the ground truth trajectories of the DiSCo data, since they are available in this context. In a real data study, the priors would be built from statistical anatomical atlases. Several synthetic data were simulated using the DWI images of the dataset by varying several parameters of the images and of the FOD estimation, in order to study the influence of those parameter on the anatomical priors informed tractography. Three different experiments were performed, as follow:Noise The DWI image were corrupted by a Rician noise resulting to a signal-to-noise-ratio (SNR)of 10,20,30,40 and 50.
Spatial down-sampling Original DWI images were down sampled with factor of 2 and 4, generating image resolution of 40x40x40, 20x20x20 and 10x10x10 voxels.
Shell selection The DiSCo DWI image gradients are distributed over 4 b-shells (b=1000, 1925,3094, 13191 s/mm2). Four mono-shell DWI images were generated by only extracting the gradients associated with the one b-shells.
Tractography was performed with two state-of-the-art algorithms, MRtrix iFOD2 7 and trekker PTT (Parallel Transport Tractography) 8, for each experiments. Overall results quality was quantified by measuring the generalized Dice score 9, based on the fibers masks and using the ground truth trajectories as reference.
Methods
Our method (Figure 1) is composed of two parts: the first one is the extraction and creation of anatomical prior from the ground-truth image and the second is the combination of the prior and FOD data.The process of prior creation is based on a modified version of Track Orientation Distribution (TOD) imaging 11. Those priors are estimated as follow. For each voxel of a fiber image, the directions of the fibers are extracted.Next, a k-means clustering is used to select the main local directions of the fibers. Then a 3D-gaussian function is fitted on each of these directions. The fitted Gaussians are then represented in spherical harmonics. Those functions are thus an image of the local mains directions of a tractograms, expressed in the same form as the diffusion FOD data. The averaging of the prior and FOD data is then performed using Riemannian geometry, as shown in 10 . In order to use the priors mainly in complex regions, the generalized fractional anisotropy (GFA) is used as a weighting factor in the average.
Results
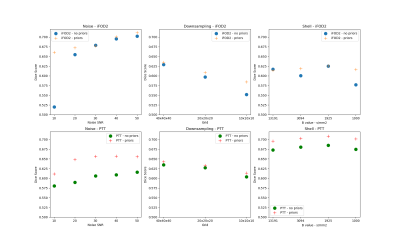
Figure 2 presents the results of the three experiments. We see that in almost all situations both algorithms perform better with anatomical prior than without. The noise experiment, in particular, shows us that, with a noise SNR below 30 dB, which is most of the time the clinical conditions,the addition of prior can lead to significant improvement in the quality of the tractograms. The down-sampling experiment demonstrates that degrading the data sampling quality will also de-grade the tractograms. The shell selection experiment shows that, depending on the algorithm,the shell used for the FOD estimation can have a small influence on tractograms quality, which can be improved with the use of anatomical prior. At last, these three experiments shows that the improvements brought by the anatomical prior are algorithm specific, which should be studied in future works.Discussion and conclusion
We have proposed a new method for creating and adding anatomical prior to tractography algorithms to enhance the overall quality of tractograms. We demonstrate that, on synthetic data,this method can lead to significant improvement, depending on conditions and the algorithm used.Future work should evaluate this method on real data as well as the prior creation process when no ground truth are available, using anatomical atlases build with tractograms segmented by expert,such as 12. It should also study the connectivity of the resulting tractograms rather than their shape. This would allow us to evaluate the influence of anatomical prior on the false positive rate of the algorithms used.Acknowledgements
No acknowledgement found.References
1. Saad Jbabdi and Heidi Johansen-Berg. Tractography: Where do we go from here? Brain Connectivity, 1(3):169–183, 2011. PMID: 22433046.
2. Derek K Jones. Challenges and limitations of quantifying brain connectivity in vivo with diffusion mri. Imaging in Medicine, 2(3):341, 2010.
3. Klaus H Maier-Hein, Peter F Neher, Jean-Christophe Houde, Marc-Alexandre Côté, Eleftherios Garyfallidis, Jidan Zhong, Maxime Chamberland, Fang-Cheng Yeh, Ying-Chia Lin, QingJi, et al. The challenge of mapping the human connectome based on diffusion tractography.Nature communications, 8(1):1–13, 2017.
4. Francois Rheault, Etienne St-Onge, Jasmeen Sidhu, Klaus Maier-Hein, Nathalie Tzourio-Mazoyer, Laurent Petit, and Maxime Descoteaux. Bundle-specific tractography with incorporated anatomical and orientational priors. NeuroImage, 186:382–398, 2019.
5. J-Donald Tournier, Fernando Calamante, and Alan Connelly. Robust determination of the fibre orientation distribution in diffusion mri: non-negativity constrained super-resolved spherical deconvolution. Neuroimage, 35(4):1459–1472, 2007.
6. Gabriel Girard, Emmanuel Caruyer, Jonathan Rafael-Patino, Marco Pizzolato, Raphaël Truffet, and Jean-Philippe Thiran. Diffusion-simulated connectivity challenge, March 2021.
7. J Donald Tournier, Fernando Calamante, Alan Connelly, et al. Improved probabilistic stream-lines tractography by 2nd order integration over fibre orientation distributions. In Proceedings of the international society for magnetic resonance in medicine, volume 1670. John Wiley &Sons, Inc. New Jersey, USA, 2010.
8. Dogu Baran Aydogan and Yonggang Shi. Parallel transport tractography. IEEE Transactions on Medical Imaging, 40(2):635–647, 2021.
9. William R Crum, Oscar Camara, and Derek LG Hill. Generalized overlap measures for evaluation and validation in medical image analysis. IEEE transactions on medical imaging,25(11):1451–1461, 2006.
10. Alvina Goh, Christophe Lenglet, Paul M. Thompson, and René Vidal. A non parametric riemannian framework for processing high angular resolution diffusion images and its applications to odf-based morphometry. NeuroImage, 56(3):1181–1201, 2011.
11. Thijs Dhollander, Louise Emsell, Wim Van Hecke, Frederik Maes, Stefan Sunaert, and PaulSuetens. Track orientation density imaging (todi) and track orientation distribution (tod)based tractography. NeuroImage, 94:312–336, 2014.
12. Jakob Wasserthal, Peter Neher, and Klaus H. Maier-Hein. Tractseg - fast and accurate whitematter tract segmentation. NeuroImage, 183:239–253, 2018.
Figures