2363
Repeatability and reproducibility of MRF-based Myelin Water Fraction maps of healthy human brains1IRCCS Stella Maris, Pisa, Italy, 2New York University, New York, NY, United States, 3University of Cambridge, Cambridge, United Kingdom, 4IMAGO7 Foundation, Pisa, Italy, 5Azienda Ospedaliero-Universitaria Pisana, Pisa, Italy, 6University of Pisa, Pisa, Italy, 7Fondazione Toscana Gabriele Monasterio, Pisa, Italy, 8GE Healthcare, Munich, Germany, 9Istituto Nazionale di Fisica Nucleare, Pisa, Italy
Synopsis
Keywords: MR Fingerprinting/Synthetic MR, MR Fingerprinting, Myelin, Reproducibility
Myelin Water Fraction (MWF) can measure White Matter (WM) myelination and integrity and can be quantified using Magnetic Resonance Fingerprinting (MRF) which allows short scan time. In this work, we assessed the repeatability and the reproducibility of MWF using 3D SSFP MRF in a traveling head study performed on healthy volunteers scanned at five different scanners from the same vendor. We computed coefficients-of-variation to estimate voxelwise variability in WM and GLM analysis to measure biases. We reported a variability of ∼3% for repeated scans at the same site and ∼5% for different sites, with an average bias of 5%.Introduction
Myelin Water Fraction (MWF) can provide quantitative information on White Matter (WM) myelination and integrity, allowing the assessment of normative brain development and the identification and longitudinal evaluation of lesions and dys/de-myelination processes. The quantification of MWF via Magnetic Resonance Fingerprinting1,2, which allows fast scans, can favor the spreading of this biomarker in clinical practice. The reproducibility of MRF-based T1, T2 and PD maps was demonstrated for both 2D and 3D SSFP sequences3–5, but the evaluation of the repeatability and reproducibility of multi-component models applied to the MRF signal is currently missing. To this aim, we performed a secondary analysis of test/retest data from a traveling head study acquired using 3D SSFP MRF in the human brain at five different 1.5T MR systems from the same vendor5.Methods
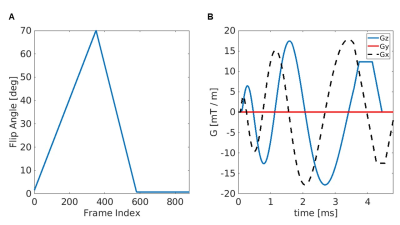
Data acquisition: Data5 were acquired on five 1.5T MR scanners from the same vendor (GE Healthcare) with different hardware and software release, using a 3D SSFP MRF6,7 sequence. The MRF acquisition trajectories used undersampled spiral projection interleaves (Figure 1) with voxel size = 1.125mm isotropic, FOV = 220mm, matrix = 200x200x200, sampling bandwidth = ±250kHz, TE/TR = 0.5/11 ms, scan duration = 8min 37s. The spirals were rewound and followed by a spoiler z-gradient achieving 4π dephasing across the voxel. The maximum gradient amplitude was 20mT/m and slew rate was 70T/m/s. No system-specific gradient delay or trajectory correction was applied. Nine healthy volunteers (age: 28-37 yo, 7 males, 2 females) participated in the study and underwent two identical separated sessions per scanner including MRF acquisition. In addition, a T1w FSPGR scan was acquired at the same spatial resolution as 3D MRF for anatomical reference. To enable a General Linear Model (GLM) analysis, each subject was scanned on at least two MR scanners.MRF reconstruction: T1, T2 and PD MRF maps were obtained by inner-product pattern between acquired and simulated signals, computed using the Extended Phase Graphs formalism8, without including B0 or B1 effects in the model. MRF-based MWF maps were obtained by removing the cerebrospinal fluid signal and matching tissue-only signal evolutions with a precomputed two-component dictionary9, i.e., Intra/Extra-cellular (IEW) and Myelin Water (MW), including chemical exchange effects (myelin fraction resolution: 0.01).
Analysis: The T1w images were warped together to create a study-specific template using ANTs. Tissue class segmentation was performed using SPM to obtain WM probabilistic masks, which were converted to binary using a threshold of 70%. The T1 map of every retest scan was registered to the corresponding test scan using ANTs. The test scans of each subject were coregistered, aligned to the corresponding T1w image and finally warped to the study-specific template. The repeatability and reproducibility of MWF were evaluated in the WM ROI after applying a 6mm-FWHM Gaussian smoothing. The Coefficient of Variation (CV) was calculated voxelwise to compare test and retest scans as well as scans performed at different sites and then averaged over the WM ROI. A GLM analysis was run in SPM to estimate the bias associated with each covariate of the experiment, i.e., test/retest, subject and site.
Results
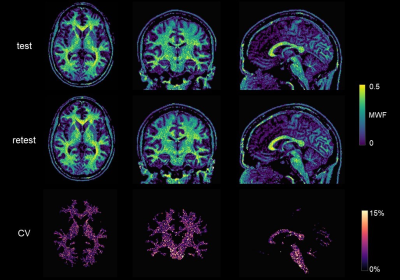
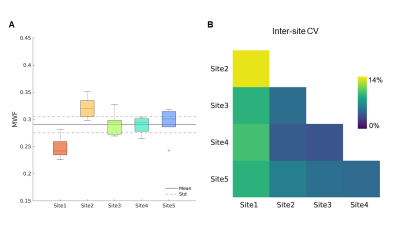
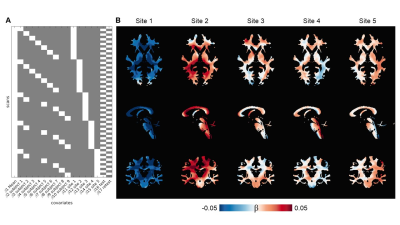
MRF-based MWF maps are shown in Figure 2. Voxelwise CV for repeated scans in WM (Figure 2) was ∼3% at all sites, except for Site 5 with CV = 6.8%. We reported MWF = 0.29 ± 0.02 on average in the whole WM (Figure 3A). The inter-scanner CV was < 5%, except when considering Site 1 which yielded an average CV of 10% (Figure 3B). The GLM approach showed that MWF biases associated with the sites ranged from 3.3% to 16.1%, with an average of 5.4% and their spatial distributions resembled those of T2 maps found in the previous study5 (Figure 4) while the test/retest bias was <1%.Discussion
In a traveling head experiment, we assessed the repeatability and the reproducibility of MWF maps obtained via 3D MRF on five different 1.5 T scanners using voxelwise CV to estimate variability and GLM analysis to estimate bias. The test-retest CV was slightly higher than the one found for T1 but comparable to that of T2 in the original study5, probably because the MWF estimate depends on both relaxation times in a non-trivial manner. Instead, the inter-site variability is similar to those of T1 and T2, likely because the contribution of the sites to the variance becomes dominant. The CV found in this study are similar to those obtained in previous works in which MWF was estimated using other techniques10,11.Conclusion
This study shows high repeatability and inter-scanner reproducibility of the MWF maps obtained using 3D SSFP MRF sequence. This reproducible metric could increase MR sensitivity and has the potential to improve the diagnosis of lesions.Acknowledgements
Support from the Italian Ministry of Health under the project BIaNCA, Pediatric Network IDEA and under the grant RC 2022 and “5 per mille” to IRCCS Fondazione Stella Maris.
Support from the Italian Ministry of Health and the Tuscany Region under the project “Ricerca Finalizzata”, Grant n. GR-2016-02361693.
Funding from the EMPIR Programme 18HLT05 QUIERO Project, co-financed by the Participating States and from the European Union’s Horizon 2020 Research and Innovation Programme.
Support from the NIHR Cambridge Biomedical Research Centre.
References
1. Ma D, Gulani V, Seiberlich N, et al. Magnetic resonance fingerprinting. Nature. 2013;495(7440):187-192. doi:10.1038/nature11971
2.Chen Y, Chen MH, Baluyot KR, Potts TM, Jimenez J, Lin W. MR fingerprinting enables quantitative measures of brain tissue relaxation times and myelin water fraction in the first five years of life. NeuroImage. 2019;186:782-793. doi:10.1016/j.neuroimage.2018.11.038
3. Jiang Y, Ma D, Keenan KE, Stupic KF, Gulani V, Griswold MA. Repeatability of magnetic resonance fingerprinting T1 and T2 estimates assessed using the ISMRM/NIST MRI system phantom. Magn Reson Med. 2017;78(4):1452-1457. doi:10.1002/mrm.26509
4. Buonincontri G, Biagi L, Retico A, et al. Multi-site repeatability and reproducibility of MR fingerprinting of the healthy brain at 1.5 and 3.0 T. NeuroImage. 2019;195:362-372. doi:10.1016/j.neuroimage.2019.03.047
5. Buonincontri G, Kurzawski JW, Kaggie JD, et al. Three dimensional MRF obtains highly repeatable and reproducible multi-parametric estimations in the healthy human brain at 1.5T and 3T. NeuroImage. 2021;226:117573. doi:10.1016/j.neuroimage.2020.117573
6. Gómez PA, Cencini M, Golbabaee M, et al. Rapid three-dimensional multiparametric MRI with quantitative transient-state imaging. Sci Rep. 2020;10(1):13769. doi:10.1038/s41598-020-70789-2
7. Kurzawski JW, Cencini M, Peretti L, et al. Retrospective rigid motion correction of three-dimensional magnetic resonance fingerprinting of the human brain. Magn Reson Med. 2020;84(5):2606-2615. doi:https://doi.org/10.1002/mrm.28301
8. Weigel M. Extended phase graphs: Dephasing, RF pulses, and echoes - pure and simple. J Magn Reson Imaging. 2015;41(2):266-295. doi:https://doi.org/10.1002/jmri.24619
9. Lancione M, Cencini M, Scaffei E, et al. Assessing white matter maturation and integrity in children using multi-component 3D-MR Fingerprinting and diffusion imaging. In: Proceedings of 30th ISMRM Annual Meeting, 2022. https://archive.ismrm.org/2022/0655.html
10. Deoni SC, Samson R, Wheeler-Kingshot CA. Intra and Inter-Site Reproducibility of Myelin Water Volume Fraction Values Derived using mcDESPOT. In: Proceedings of 17th ISMRM Annual Meeting, 2009:1
11. Meyers SM, Vavasour IM, Mädler B, et al. Multicenter measurements of myelin water fraction and geometric mean T2: Intra- and intersite reproducibility. J Magn Reson Imaging. 2013;38(6):1445-1453. doi:10.1002/jmri.24106
Figures