2360
Latent Diffusion Models Allow Generation of Synthetic Breast MRI DCE-MIPs
Lukas Folle1, Lorenz Kapsner2,3, Andreas Maier1, Michael Uder2, Sabine Ohlmeyer2, and Sebastian Bickelhaupt2
1Computer Science, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, 2Institute of Radiology, Universitätklinikum Erlangen-Nürnberg, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, 3Universitätklinikum Erlangen-Nürnberg, Friedrich-Alexander-Universität Erlangen-Nürnberg, Medizinisches Zentrum für Informations- und Kommunikationstechnik, Erlangen, Germany
1Computer Science, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, 2Institute of Radiology, Universitätklinikum Erlangen-Nürnberg, Friedrich-Alexander-Universität Erlangen-Nürnberg, Erlangen, Germany, 3Universitätklinikum Erlangen-Nürnberg, Friedrich-Alexander-Universität Erlangen-Nürnberg, Medizinisches Zentrum für Informations- und Kommunikationstechnik, Erlangen, Germany
Synopsis
Keywords: MR Fingerprinting/Synthetic MR, Breast
The training of neural networks for classification or segmentation of medical images requires large amounts of training data. Sharing of these datasets is commonly difficult due to legislation and privacy constraints of medical data. In this work, we demonstrate the utility of latent diffusion models that allow the generation of synthetic samples of dynamic contrast-enhanced breast MRI-derived maximum intensity projections of subtraction series. Whilst the image quality of the generated data is high as demonstrated by a radiologist evaluation, further steps are envisioned to derive specific compounds of data, e.g., BI-RADS, FGT, or BPE classes.Introduction
An increasing number of works use deep learning to help radiologists in classifying, segmenting, and measuring disease entities in medical images [3, 4]. A key resource to train such methods among necessary computing resources is a large dataset that covers the different manifestations of the entity in the images for the task at hand. For instance, to train a neural network for BI-RADS classification on breast MRI that generalizes well, different patient anatomies as well as a balanced distribution of the different classes is necessary. With recent updates in privacy regulations, the sharing of large datasets becomes increasingly difficult [5].Generative neural networks aim at learning the distribution of a given dataset and allow sampling of new cases. Compared to earlier methods, latent diffusion models (LDM) have recently demonstrated to produce realistic images of outstanding quality [1,2]. In this feasibility work, we use latent diffusion models to overcome the above-mentioned privacy constraints by training a neural network that generates synthetic MRI DCE MIPs of the breast. This way, instead of sharing real patient data, the model could be shared, which enables the generation of arbitrarily many cases.
Methods
The retrospective study was approved by the ethics committee of the Friedrich-Alexander-University Erlangen-Nürnberg and included clinically indicated breast MRI examinations of women, which were acquired with a full diagnostic protocol at the Institute of Radiology of the University Hospital Erlangen (UHE). The dataset is partly overlapping with a previously published study sample [6].Latent diffusion models consist of two parts: An autoencoder, which reduces the dimensionality of the data and transforms the data into the latent space, and the diffusion model itself. In this work, we closely follow the work by Rombach et al. [1]. The autoencoder is based on the VQ-VAE model and consists of three stages with 32, 64, and 128 channels, respectively. The diffusion model is based on a U-Net with 32, 64, and 128 channels for each stage. It is trained without conditioning and with a batch size of 16, L1 loss function, a learning rate of 1e-6, and 1000 diffusion steps. The generation of new samples is performed using 200 DDIM steps.
To evaluate the quality of the generated images, a reading by a trained radiologist was performed. In a blinded manner, 100 images of the dataset and 100 images generated by the LDM were shown. The reader was then asked to grade how realistic the images look on a scale from 0 to 4, where 0 corresponds to an unrealistic image, 1 to a slightly unrealistic image, 2 to an indeterminate image, 3 to a quite realistic image, and 4 to a fully realistic image.
Results
The dataset consisted of 2840 breast MRI examinations of 1970 individuals. The median age at first examination in the dataset was 50 years (IQR: 42 to 59). Images produced by the LDM are shown in Figure 1. The LDM-derived data achieved to represent different anatomic conditions including post-surgical reflecting unilateral (d) mastectomy, different quality including artifacts reflecting realistic shapes and regions in which artifacts are commonly noticed, and cases with and without lesions (a, c, e). The distribution of the real and synthetic cases across the Likert scores is visualized in Figure 2. As expected, the real images achieved very high scores with a maximum of 89 cases for score 4, while the cases for the synthetic images are spread more evenly across the Likert scores, however, the highest number of cases (32) is also present within the group of fully realistic images (score 4). During the evaluation it was apparent that some features like anatomical patterns of blood vessels seemed yet to be challenging to be reliably realistically represented in the generated MIPs (e.g., visible by a net-like blood vessel architecture in a).Discussion
In this work, we demonstrated the feasibility of LDMs to generate MIPs of contrast-enhanced breast MRI subtraction series. Possible applications of this work include the generation of large datasets for the training of neural networks for various tasks, but also education of medical students.This initial study's images are already of high quality, as assessed by a radiologist using visual assessments and a Likert-like scale in a blinded reading. However, it is apparent that some anatomical features, especially regarding the vessel architecture within the breast, sometimes do not reflect real anatomy, indicating that the LDM might yet be suffering from an insufficient sample size.
To be able to use the synthetic images for training of classification models, a conditioning on class labels is necessary. Herein, a special focus might be placed on deriving the most relevant clinical classes in breast MRI as a condition to be selected, which might include BI-RADS scores, lesion types and sizes, and BPE and FGT.
Conclusion
Latent diffusion models are able to generate realistic breast MRI subtraction MIPs reflecting different clinically relevant conditions. Future work on improving the model and providing a selectable conditioning as well as research on data privacy of the generated data is necessary.Acknowledgements
No acknowledgement found.References
- Rombach, R., Blattmann, A., Lorenz, D., Esser, P., & Ommer, B. (2022). High-Resolution Image Synthesis with Latent Diffusion Models. 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 10674-10685.
- Martin Arjovsky, & Leon Bottou (2017). Towards Principled Methods for Training Generative Adversarial Networks. In International Conference on Learning Representations.
- Schelb, P., Kohl, S., Radtke, J., Wiesenfarth, M., Kickingereder, P., Bickelhaupt, S., Kuder, T., Stenzinger, A., Hohenfellner, M., Schlemmer, H.P., Maier-Hein, K., & Bonekamp, D. (2019). Classification of Cancer at Prostate MRI: Deep Learning versus Clinical PI-RADS Assessment. Radiology, 293(3), 607-617.
- Akkus, Z., Galimzianova, A., Hoogi, A. et al. Deep Learning for Brain MRI Segmentation: State of the Art and Future Directions. J Digit Imaging 30, 449–459 (2017). https://doi.org/10.1007/s10278-017-9983-4
- McCall, B. (2018). What does the GDPR mean for the medical community? The Lancet, 391(10127), 1249–1250. doi:10.1016/S0140-6736(18)30739-6
- Kapsner, Lorenz A., Sabine Ohlmeyer, Lukas Folle, Frederik B. Laun, Armin M. Nagel, Andrzej Liebert, Hannes Schreiter, et al. 2022. “Automated Artifact Detection in Abbreviated Dynamic Contrast-Enhanced (DCE) MRI-derived Maximum Intensity Projections (MIPs) of the Breast.” European Radiology. https://doi.org/10.1007/s00330-022-08626-5.
Figures
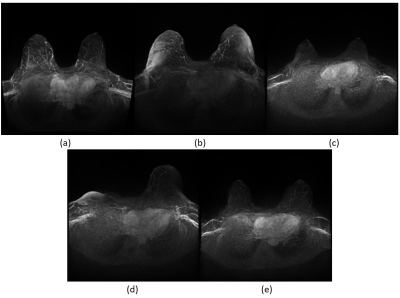
Images generated by
the latent diffusion model trained on breast MRI MIPs. The predictions cover
typical anatomical conditions both reflecting pre- and postsurgical cases
(e.g., unilateral mastectomy in (d), image artifacts, (b, d), background
parenchymal enhancement patterns, and lesions (a).
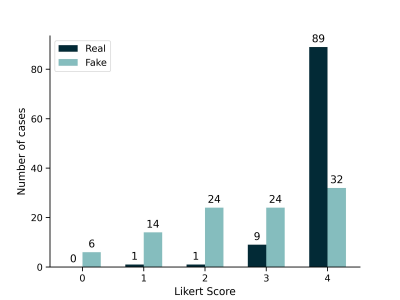
Realisticness of the
images. The bar plot shows the results of the blinded reading of 100 real and
100 synthetic images by the radiologist, stratified by real and synthetic
images. The grading was performed on a Likert scale: 0 corresponds to an
unrealistic image, 1 to a slightly unrealistic image, 2 to an indeterminate
image, 3 to a quite realistic image, and 4 to a fully realistic image.
DOI: https://doi.org/10.58530/2023/2360